Introduction
Esophageal Cancer (EC) is the eighth most incidence tumor and
is the sixth leading cause of cancer-related mortality worldwide,
indicating a high EC-associated lethality [1]. The areas with the
highest EC incidence comprise Asia, Southern and Eastern Africa,
the United Kingdom, and some countries of South America, such
as Brazil [2]. This tumor is classified into two main histological
subtypes, Esophageal Adenocarcinoma (EAC) and Esophageal
Squamous Cell Carcinoma (ESCC), the latter accounting for more
than 90% of all cases worldwide [2].
Several studies have applied Next-Generation Sequencing
(NGS) to describe the main molecular alterations in ESCC. Recently, the Cancer Genome Atlas Consortium (TCGA) published the integrated genomic characterization of EC, showing both mutations
and copy number variations affecting cell cycle regulators in this
tumor. In concordance with other studies, this study also observed that TP53 is mutated in 70-90% of ESCC samples [3-6]. TP53
encodes the p53 protein, that can bind to specific promoter sequences and transactivate a wide range of genes such as CDKN1A
(p21CIP1) and GADD45Aand has a central role in a very complex
network involved in cell cycle regulation [7]. For example, p21inhibits the activity of cyclin-CDK2 or CDK4 complexes and thus plays
a role as a p53 effect or in cell cycle control [8-10]. In addition,
GADD45A is often induced by p53 in response to DNA damage
and other stress signals, triggering cell growth arrest, DNA repair,
and apoptosis [7,11].
Regarding copy number variations, CGH (Comparative genomic
hybridization) analysis has identified CDKN2A deletion in 20% to
76% of ESCC cases evaluated [6,12]. On the other hand, recently,
in a meta-analysis, which included data from 41 case-control studies, including Asian, Caucasian, and African patients, CDKN2A
promoter methylation was significantly higher in EC samples than
in healthy controls [13]. CDKN2A locus codes for two proteins,
p14 and p16, which are involved in the p53-dependent regulation of cell cycle progression [14]. Altogether, these studies have
shown that cell cycle homeostasis disruption is fundamental to
promoting and progressing ESCC carcinogenesis, with a pivotal
role of TP53 and its molecular partners. However, the impact of
the differential expression of these genes onpatients’ prognosis is
not yet clear and overall controversial.
Therefore, this study aimed to (i) evaluate the gene expression
profile of key cell cycle regulators related to p53 function in ESCC
and (ii) to determine their potential as prognosis biomarkers.
Materials and methods
Human Samples
Seventy-five matched biopsies were collected from patients
diagnosed with ESCC (tumor tissue and non-tumor surrounding
mucosa, collected 4 inches from the tumor border) who underwent surgery or endoscopy between 2000-2007. In total, 46 patients from the Southeast region of Brazil (Rio de Janeiro and São
Paulo) were included in this study: Three from Hospital Universitário Pedro Ernesto, UERJ, Rio de Janeiro; 22 from INCA, Rio de Janeiro; and 21 from Surgery Department, UNICAMP, São Paulo.
The remaining 29 samples were collected in Hospital das Clínicas dePorto Alegre, HCPA/UFRGS, Rio Grande do Sul, in the South
of Brazil. Patients enrolled in this study had not undergone prior
chemotherapy or radiotherapy. Patients’ habits regarding smoking and alcohol consumption, socio-demographic characteristics, tumor differentiation, and esophageal location were collected by a standardized questionnaire and from hospital records.
The institutions’ Ethics Committees approved this study, and all
procedures followed the 1964 Helsinki declaration and its later
amendments or comparable ethical standards. All patients signed
written informed consent.
RNA extraction, reverse transcription (RT) and quantitative
PCR (qPCR)
Total RNA was extracted from biopsies using TRIzol® (Invitrogen, USA), following the protocol described by the manufacturer. Next, cDNA was synthesized using Super Script™ First-Strand
Synthesis System and random hexamers, according to the manufacturer’s instruction. qPCR was used to evaluate p14ARF,
p16INK4a, p21CIP1, TP53, and GADD45A mRNA expression (Supplementary Table S1). GAPDH and ACTB(Supplementary Table S1)
were used as housekeeping genes. The number of samples analyzed for each gene varied due to RNA availability. qPCR was performed with the ABI 7700 detection system (Applied Biosystems,
USA) as previously described [15].
DNA extraction and bisulfite treatment: p14ARF and p16IN-K4a mRNA expression levels were categorized into low (log2 fold-change<-1), no change (-1≤log2 fold-change≤1) and high (log2 fold-change>1). Next, we randomly selected 10 ESCC samples (tumor
and non-tumor surrounding mucosa) from each category (high,
no change, and low) to perform DNA extraction, followed by bisulfite treatment and pyrosequencing. DNA was extracted from
thirty frozen ESCC sample pairs by SDS/proteinase K protocol [16].
Then, 1.0 μg of DNA was treated using the EpiTect® Bisulfite kit
(Qiagen, Germany) according to the manufacturer’s instructions
to convert unmethylated cytosine residues to uracil, leaving the
methylated cytosines unchanged.
Pyrosequencing
The methylation status of ten selected CpG sites in p14ARF and
p16INK4a promoters was analyzed by pyrosequencing. Bisulfite-treated DNA (25 ng) was used to amplify the regions of interest
with primers designed with the PSQ TM24MA System software
(Qiagen, Germany) (Supplementary Table S1). Taq platinum DNA
polymerase (Invitrogen, USA) was used for PCR reaction following
manufactures protocol. Amplification was performed with 5 min
at 95oC, followed by 40 cycles of 30 sec at 95oC, 45 sec at the
specific temperature for each pair of primers, and 30 sec at 72oC,
followed by one hold at 72oC for 10 min. Pyrosequencing was performed according to the manufacturer’s protocol (Qiagen, Germany). The target CpGs were evaluated by converting the resulting pyrograms into numerical values for peak heights and calculating the mean of all CpG sites analyzed at a given gene promoter.
Samples that showed low-quality peaks were excluded from the
analysis.
Statistical analysis
The Wilcoxon matched pairs test or Kruskal Wallis test was
used to assess mRNA expression or methylation percentage differences between tumors and non-tumor surrounding mucosa
using GraphPad 5.0 software (GraphPad Software, Inc., San Diego,
CA, USA). Differences were considered statistically significant
when p<0.05. Overall survival was analyzed 24 months after diagnosis. Patients who were alive at the end of the follow-up period
were censored. For those who were lost to follow-up, the date
of the last information obtained was considered for purposes of
censorship.
Furthermore, the impact of mRNA expression of each gene on
overall survival was evaluated and this molecular variable was categorized into tertiles. Patients falling in the lower and middle tertiles were grouped and compared to patients falling in the highest
tertile. The Kaplan-Meier method was used to assess univariate
survival, while statistical significance between groups was calculated with the log-rank test, assuming a statistical significance
level of 5%. The Cox proportional hazards regression model was
used for univariate and multivariate analyses to explore the relationship between the mRNA expression of each gene analyzed
and the prognostic value for survival. Variables that showed
p<0.20, age, and tumor stage were used to adjust the association
between mRNA expression and global survival. All data were analyzed using the statistical package SPSS for Windows 20.0.
Results
Clinicopathological characteristics
The median age of patients included in this study was 56 years
[34-83], and most were male (82.7%), alcohol drinkers (88.0%),
and tobacco smokers (86.6%), with 81.3% presenting both habits.
Most tumors were in the middle or distal thirds of the esophagus
(66.6%), showed moderate or well differentiation status (61.4%)
and were diagnosed in the advanced clinical stage (66.7%) (Table
1).
Gene expression profile
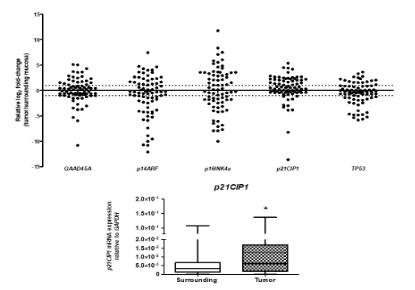
We evaluated the expression of GADD45A, p14ARF, p16INK4a,
p21CIP1, and TP53 in ESCC and non-tumor surrounding mucosa, and tumors samples showed p21CIP1overexpression (fold-change: 2,0; p = 0.0011), while the other genes showed no statistically significant expression differences (Figure 1B). Furthermore,
it is noteworthy that ESCC samples presented heterogeneity in
the p14ARF and p16INK4a expression compared to matched non-tumor adjacent mucosa (fold-change), which was not observed in
the other analyzed genes (Figure 1A).
Table 1: Clinicopathological and socio-demographic data of ESCC
patients.
| SOCIO-DEMOGRAPHIC DATA |
TOTAL(n#=75) |
AGE |
| Median (min-max) |
56.0(34-83) |
Gender |
| Men |
62(82.7%) |
| Womenen |
13(17.3%) |
Origin |
| Southeast |
46(61.3%) |
| South |
29(38.7%) |
Alcohol Consumption |
| Never |
7(9.3%) |
| Ever |
66(88.0%) |
| Missing |
2(2.7%) |
Tobacco Consumption |
| Never |
8(10.7%) |
| Ever |
65(86.6%) |
| Missing |
2(2.7%) |
CLINICAL DATA |
| Tumor Location |
| Proximal esophagus |
5(6.7%) |
| Middle esophagus |
33(44.0%) |
| Distal esophagus |
17(22.6%) |
| More than one region affected |
15(20.0%) |
| Missing |
5(6.7%) |
Tumor Differentiation |
| Well and Moderately |
46(61.4%) |
| Poorly and Undifferentiated |
19(25.3%) |
| Missing |
10(13.3%) |
Stage |
| I+II |
21(28.0%) |
| III+IV |
50(66.7%) |
| Missing |
4(5.3%) |
T stage |
| T1+T2 |
6(8.0%) |
| T3+T4 |
61(81.3%) |
| Missing |
8(10.7%) |
Lymph node invasion |
| No |
34(45.3%) |
| Yes |
33(44.0%) |
| Missing |
8(10.7%) |
Survival (months) |
| Median (min-max) |
12(1-99) |
#number of patients
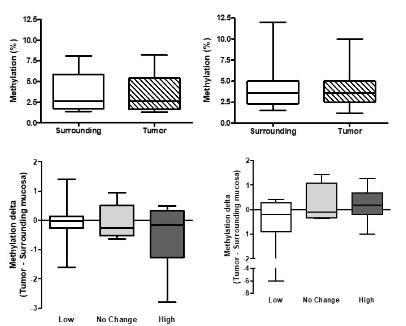
p16INK4a and p14ARF DNA methylation and expression analysis
The methylation status of p14ARF and p16INK4a promoters
was evaluated in to address its correlation with gene expression as a possible explanation for the heterogeneity observed in
ESCC samples. First, pyrosequencing revealed that p16INK4a and
p14ARF promoters present similar methylation levels in tumors
and surrounding mucosa without significant differences in the
mean methylation of the 10 CpG sites analyzed (Figure 2 A & B) or
in CpG sites analyzed individually (data not shown). Next, samples
were subcategorized into three groups according to the expression levels of p14ARF and p16INK4a in ESCC in comparison to matched non-tumor adjacent mucosa. So, we assessed whether there
would be an association between the delta methylation (tumor-surrounding mucosa) and the mRNA expression levels according
to the three subcategorized groups described above, but no statistically significant difference was observed (Figure 2 C & D). Finally,
we investigated the association between p14ARF and p16INK4a
up and down regulation and clinical or socio-demographic parameters, including 2-year overall survival. We found no significant
association between their expression and the evaluated parameters (Supporting information Table S2).
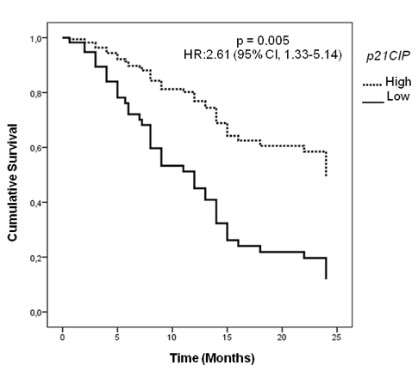
Impact of differential p21CIP1 expression on ESCC patient’s
overall survival
Following the observed p21CIP1 deregulation in ESCC, we
evaluated the association between its findings and clinical or
socio-demographic parameters, and no significant associations
between p21CIP1 expression and these parameters were found
(Supporting information Table S3). Next, we investigated the impact of p21CIP1 over expression on ESCC overall survival. Multivariate analysis revealed that patients with low p21CIP1 expression
presented a median 2-year survival rate of 12%, the patients with
high p21CIPI levels group did not reach the 50% survival mark in
the evaluated period, indicating that p21CIP1 expression is an independent prognostic factor in ESCC Brazilian patients HR: 2.61
(95% CI, 1.33-5.14); p = 0.005) (Figure 3).
Discussion
In the present study, we determined the mRNA expression and
methylation changes in genes that encode components of the
p53-dependent cell cycle regulation pathway in paired samples of
ESCC and non-tumor adjacent mucosa. We found high heterogeneity in p14ARF and p16INK4a expression; however, the methylation status of the promoter region of these genes was not correlated with this phenomenon. Interestingly, among all investigated
genes, only p21CIP1(CDKN1A) was found to be overexpressed in
ESCC and showed to be an independent prognostic factor.
In this study, we did not observe associations between p14ARF
and p16INK4a expression and promoter methylation status in
ESCC samples. This data agrees with our previous results, showing
no differences in the methylation levels of CDKN2A locus in ESCC
samples compared with non-tumor surrounding mucosa and esophageal mucosa from healthy individuals [17]. However, other
studies have detected promoter hyperm ethylation in these genes
in ESCC [12,13,18]. This apparent discrepancy could be explained
by the different methods used to analyze the methylation profile,
like methylation-specific PCR (MSP), or it could reflect differences
between the populations studied. The etiological factors associated with ESCC development vary worldwide and could impact the
expression or activity of cell cycle regulators, especially p14ARF
and p16INK4a, by distinct molecular mechanisms, including homozygous deletions, intragenic mutations, and complex regulatory nets of non-coding RNAs [6,17,19-21]. Recently, the TCGA
report that CDKN2A inactivation is a common trait for ESCC (76%)
and is usually associated with deep gene deletion [6]. Therefore,
although different inactivating molecular alterations have been
reported in the CDKN2A locus, their impact on gene and protein
expression in ESCC needs to be further evaluation.
Among the cell cycle and p53-regulated genes evaluated in
this study, only p21CIP1was upregulated in ESCC compared to non-tumor adjacent mucosa. Furthermore, ESCC patients with
low p21CIP1 expression had a shorter 2-year overall survival. The
impact of p21 expression on patients’ prognosis has been previously reported for several tumors, including colorectal, bladder,
and gastric cancer [22-24]. Furthermore, such impact seems to
be dependent on the age at diagnosis. For example, the loss of
this tumor suppressor was associated with a worse prognosis in
younger colorectal cancer patients (HR 4.09, 95% CI, 1.13-14.9)
but showed the opposite trend in older individuals (HR 0.37; 95% CI, 0.24-0.59) [22]. Meanwhile, in gastric cancer, increased levels
of p21 protein were associated with improved overall survival in
older patients [24]. Thus, we did not observe any significant association between age, p21CIP1 expression, and ESCC patients’
overall survival.
In ESCC, previous studies have shown discordant associations
between p21 expression and 5-year overall survival, with reports
suggesting a lack of association [25,26] and others showing a negative association between p21 over expression and survival of
patients [27,28]. Nonetheless, different authors have shown that
the over expression of this protein has a positive impact on prognosis [26,29,30], similar to our findings. Therefore, the impact of
p21 mRNA and protein levels on overall survival should be further
explored in ESCC since it might be dependent on patients’ age and
the studied population.
Unfortunately, there are no drugs in clinical practice that target p21 [31]. However, drugs tested in ESCC cell cultures, such
as Obatoclax (an inhibitor of Bcl-2 family members) and diallyl
disulfide (an organosulfur compound derived from garlic), have
shown as apparent antitumor effect on these cells [32,33]. Furthermore, treatment with these drugs resulted in cell cycle arrest,
reduced cell viability, induction of apoptosis, and increased p21
expression. Thus, studies that assess the potential of p21 as a therapeutic target could improve the prognosis of patients with low
expression of this protein.
TP53 expression did not show differences between ESCC and
non-tumor surrounding mucosa. Since p21CIP1 regulation is mediated at least in part by TP53, we evaluated the possible correlation between p21CIP1 and TP53 expression, observing a positive
correlation between their mRNA levels (r = 0.55; p <0.0001, data
not shown). This moderate correlation may indicate that other
factors could be involved in p21CIP1 regulation in ESCC, such
as TP53 mutational status. Although we did not perform mutation screening in the present study, no correlation was observed
between p21CIP1expression and TP53 mutational status in the
ESCC TCGA dataset (p=0.2015, data not shown). This could be explained by the plethora of p53-independent pathways and transcription factors capable of inducing p21CIP1 expressions, such as
the Ras-Raf-Mapk oncogenic pathway, and major transcriptional
regulators, such as SP1 and STAT [34,35].
Although the prognostic value of p21 expression has been previously explored in cancer, it is essential to notice that this is the
first study to address its association with ESCC survival by evaluating mRNA expression by qPCR, a quantitative method. Other
studies in ESCC and different tumors have focused on p21 protein
expression using IHC, a semi-quantitative method. Besides, these
studies used different criteria to evaluate and stratify the immune
staining pattern, with controversial results. Therefore, we suggest
that future studies should be performed to evaluate the impact of
p21CIP1 mRNA levels in ESCC by quantitative methods to better
stratify prognosis and potentially intervene to improve patients’
survival.
Conclusion
In conclusion, we showed an up regulation of p21CIP1 in ESCC,
although low p21CIP1 mRNA levels independently predict a poorer 2-year overall survival of ESCC patients.
Declarations
Conflicts of interest: The authors declare that they have no
conflicts of interest.
Funding sources: Research was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil)
and Fundação de Amparo è Pesquisa do Estado do Rio de Janeiro
(FAPERJ, Brazil).
Acknowledgments: The authors would like to thank the Endoscopy and Surgery Section of all institutions (INCA, UERJ, HCPA
and UNICAMP) that took part in this studyfor their support in the
collection samples and the National Tumor Bank (BNT) of INCA for
its support in the processing of samples.
References
- Ferlay J, Ervik M, Lam F, Colombet M, Mery L, et al. Global Cancer
Observatory: Cancer Today. Lyon, France: International Agency for
Research on Cancer. 2022.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and
mortality worldwide for 36 cancers in 185 countries. CA Cancer J
Clin. 2018; 68: 394-424.
- de Souza-Santos PT, Soares Lima SC, Nicolau-Neto M, Boroni N,
Meireles Da Costa L, et al. Mutations, Differential Gene Expression,
and Chimeric Transcripts in Esophageal Squamous Cell Carcinoma
Show High Heterogeneity, Transl Oncol. 2018; 11: 1283-1291.
- Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, et al. Genomic
Landscape of Esophageal Squamous Cell Carcinoma in a Japanese
Population, Gastroenterology. 2016: 150: 1171-1182.
- Gao YB, Chen ZL, Li JG, da Hu X, Shi XJ, et al. Genetic landscape of
esophageal squamous cell carcinoma. Nat Genet. 2014; 46: 1097-1102.
- Kim J, Bowlby R, Mungall AJ, Robertson AG, Odze RD, et al. Integrated genomic characterization of oesophageal carcinoma, Nature.
2017; 541: 169-175.
- El-Deiry WS. Regulation of p53 downstream genes, Semin Cancer
Biol. 1998; 8: 345-357.
- Yue X, Gregory H, Hui Z, David C, Kobayashi R, et al. P21 Is a Universal Inhibitor of Cyclin Kinases, Nature. 1993; 366: 701-704.
- He G, Siddik ZH, Huang Z, Wang R, Koomen J, et al. Induction of
p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2
activities.2005; 2929-2943.
- El-Deiry WS. p21 (WAF1) Mediates Cell-Cycle Inhibition, Relevant
to Cancer Suppression and Therapy. Cancer Res. 2016; 76: 5189-5191.
- Tamura RE, de Vasconcellos JF, Sarkar D, Libermann TA, Fisher PB,
et al. GADD45 proteins: central players in tumorigenesis. Curr Mol
Med. 2012; 12: 634-651.
- Ishii T, Murakami J, Notohara K, Cullings HM, Sasamoto H, et al.
Oesophageal squamous cell carcinoma may develop within a
background of accumulating DNA methylation in normal and dysplastic mucosa, Gut. 2007; 56: 13-19.
- Zhou C, Li J, Li Q. CDKN2A methylation in esophageal cancer: a
meta-analysis, Oncotarget. 2017; 8: 50071-50083.
- Joerger AC, Fersht AR. The p53 Pathway: Origins, Inactivation in
Cancer, and Emerging Therapeutic Approaches, Annu Rev Biochem. 2016; 85: 375-404.
- de A Simão T, Souza-Santos PT, de Oliveira DSL, Bernardo V, Lima
SCS, et al. Quantitative evaluation of SPRR3 expression in esophageal squamous cell carcinoma by qPCR and its potential use as a
biomarker. Exp Mol Pathol. 2011; 91.
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual,
3th edn, Cold Spring Harbor Laboratory, New York, 2001.
- Lima SCS, Hernandez-Vargas H, Simão T, Durand G, Kruel CDP, et al.
Identification of a DNA methylome signature of esophageal squamous cell carcinoma and potential epigenetic biomarkers. Epigenetics. 2011; 6: 1217-1227.
- Peng X, Xue H, Lü L, Shi P, Wang J, et al. Accumulated promoter
methylation as a potential biomarker for esophageal cancer, Oncotarget. 2017; 8.
- Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, et al. Hot air mediates
hepatocarcinogenesis through suppressing miRNA-218 expression
and activating P14 and P16 signaling, J Hepatol. 2015; 63: 886-895.
- Zhou C, Li J, Li Q. CDKN2A methylation in esophageal cancer: a
meta-analysis, Oncotarget. 2017; 8.
- Mohseny AB, Tieken C, van der Velden PA, Szuhai K, de Andrea C,
et al. Cleton-Jansen, Small deletions but not methylation underlie CDKN2A/p16 loss of expression in conventional osteosarcoma,
Genes Chromosomes Cancer. 2010; 49: 1095-1103.
- Ogino S, Nosho K, Shima K, Baba Y, Irahara N, et al. p21 expression in colon cancer and modifying effects of patient age and body
mass index on prognosis, Cancer Epidemiology Biomarkers and
Prevention. 2009.
- Shariat SF, Tokunaga H, Zhou J, Kim J, Ayala GE, et al. p53, p21, pRB,
and p16 expression predict clinical outcome in cystectomy with
bladder cancer, Journal of Clinical Oncology. 2004.
- Gomyo Y, Ikeda M, Osaki M, Tatebe S, Tsujitani S, et al. Expression
of p21 (waf1/cip1/sdi1), but not p53 protein, is a factor in the survival of patients with advanced gastric carcinoma, Cancer. 1997;
79: 2067-2072.
- Güner D, Sturm I, Hemmati P, Hermann S, Hauptmann S, et al. Multigene analysis of Rb pathway and apoptosis control in esophageal
squamous cell carcinoma identifies patients with good prognosis,
Int J Cancer. 2003; 103: 445-454.
- Nita ME, Nagawa H, Tominaga O, Tsuno N, Hatano K, et al.
p21Waf1/Cip1 expression is a prognostic marker in curatively
resected esophageal squamous cell carcinoma, but not p27Kip1,
p53, or Rb. Ann Surg Oncol. 1999; 6: 481-488
- Goan YG, Hsu HK, Chang HC, Chou YP, Chiang KH, et al. Deregulated
p21 WAF1 over expression impacts survival of surgically resected
esophageal squamous cell carcinoma patients, Annals of Thoracic
Surgery. 2005; 80: 1007-1016.
- Taghavi N. Association of p53/p21 expression with cigarette smoking and prognosis in esophageal squamous cell carcinoma patients. World J Gastroenterol. 2010; 16: 4958.
- Lin Y, Shen LY, Fu H, Dong B, Yang HL, et al. P21, COX-2, and E-cadherin are potential prognostic factors for esophageal squamous
cell carcinoma, Diseases of the Esophagus. 2017; 30 1-10.
- Guo R, Ma Y, Zhao M, Zhang W, An G, et al. Polymorphism
rs2395655 affects LEDGF/p75 binding activity and p21WAF1/CIP1
gene expression in esophageal squamous cell carcinoma, Cancer
Med. 2019; 1-12.
- Wang L, Han H, Dong L, Wang Z, Qin Y. Function of p21 and its therapeutic effects in esophageal cancer (Review), Oncol Lett. 2020;
2: 136.
- Li J, Xu J, Li Z. Obatoclax, the pan-Bcl-2 inhibitor sensitizes hepatocellular carcinoma cells to promote the anti-tumor efficacy in combination with immune checkpoint blockade, Transl Oncol. 2021;
14: 101116.
- YIN X, ZHANG R, FENG C, ZHANG J, LIU D, et al. Diallyl disulfide
induces G2/M arrest and promotes apoptosis through the p53/
p21 and MEK-ERK pathways in human esophageal squamous cell
carcinoma, Oncol Rep. 2014; 32: 1748-1756.
- Gartel AL, Tyner AL. Transcriptional Regulation of the p21(WAF1/
CIP1). Gene, Exp Cell Res. 1999; 246: 280-289.
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple
activities, Nat Rev Cancer. 2009; 9: 400-414.
Supplementary table S1: Specific sequences of forward and reverse primers used in RT-qPCR or pyrosequencing.
| GENE |
PRIMERS |
PRODUCT SIZE(bp#) |
ASSAY |
REFERENCES |
| p14ARF |
F:5'TCCTAGAAGACCAGGTCATGATG 3' |
194 |
qPCR |
Designed by the authors |
| R:5'ACCACCAGCGTGTCCAGGAA 3' |
| p16INK4a |
F:5'CAACGCACCGAATAGTTACG 3' |
171 |
qPCR |
Haller et al.,2005 |
| R:5'ACCAGCGTGTCCAGGAAG 3' |
| p21CIP1 |
F:5'ACCTGTCACTGTCTTGTACCCTTGT 3' |
121 |
qPCR |
Designed by the authors |
| R:5'TGGTAGAAATCTGTCATGCTGGT 3' |
| TP53 |
F:5'TAAGCGAGCACTGCCCAACA 3' |
96 |
qPCR |
Designed by the authors |
| R:5'TCACGCCCACGGATCTGAAG 3' |
| p21CIP1 |
F:5'ACCTGTCACTGTCTTGTACCCTTGT 3' |
121 |
qPCR |
Designed by the authors |
| R:5'TGGTAGAAATCTGTCATGCTGGT 3' |
| TP53 |
F:5'TAAGCGAGCACTGCCCAACA 3' |
96 |
qPCR |
Designed by the authors |
| R:5'TCACGCCCACGGATCTGAAG 3' |
| GADD45A |
F:5'AGAGCAGAAGACCGAAAGGATG 3' |
123 |
qPCR |
Designed by the authors |
| R:5'TCGACGTTGAGCAGCTTGGC 3' |
| GAPDH |
F:5'CAACAGCCTCAAGATCATCAGCAA 3' |
123 |
qPCR |
Designed by the authors |
| R:5'AGTGATGGCATGGACTGTGGTCAT 3' |
| β-actin |
F:5'CCAGATCATGTTTGAGACCTT 3' |
107 |
qPCR |
Designed by the authors |
| R:5'CGGAGTCCATCACGATGCCAG 3' |
| p16INK4a |
F:5'GGGTGGGGGAGTATATAGGG 3' |
163 |
Pyrosequecing |
Designed by the authors |
| R:5'biotin TCCCACCCCAACCTCCAAAATCT 3' |
| S: 5' AGGAGGGAGGGAGAGG 3' |
| p14ARF |
F:5'GGGATATGGAGGGGGAGAT 3' |
183 |
Pyrosequecing |
Designed by the authors |
| R:5'biotin TCCCCTCCCCTACTAACC 3' |
| S: 5' GAGAAAGTAAGTAGAGGAGTTAGG 3' |
#base pair; F: forward; R: reverse.
Supplementary table S2: Association between p14ARF and p16INK4a mRNA expression and clinical and socio-demographic parameters.
| SOCIO-DEMOGRAPHIC DATA |
P14ARF |
P16INK4a |
|
TOTAL (n#=54) |
High (n#=25) |
Low (n#=29) |
p value*
| TOTAL (n#=63) |
High (n#=34) |
Low (n#=29) |
p value* |
| Age |
| Median (min-max) |
56.0 (34-83) |
56.0 (34-76) |
56.0 (47-83) |
0.9515 |
56.0 (34-83) |
55.5 (40-75) |
57.0(34-83) |
0.6586 |
| Gender |
|
|
|
|
|
|
|
|
Men |
43 (79.6%) |
21 (84.0%) |
22 (75.8%) |
0.5166 |
52 (82.5%) |
29 (85.3%) |
(79.3%) |
0.7406 |
| women |
11 (20.4%) |
4 (16.0%) |
7 (24.2%) |
11 (17.5%) |
5 (14.7%) |
6 (20.7%) |
|
| Origin |
|
|
|
|
|
|
|
|
| Southeast |
34 (63.0) |
18 (72.0%) |
16 (55.0%) |
0.2628 |
38 (60.3%) |
23 (67.6%) |
15 (51.7%) |
0.3015 |
| South |
20 (37.0%) |
7 (28.0%) |
13 (45.0%) |
|
25 (39.7%) |
11 (32.4%) |
14 (48.3%) |
|
| Alcohol Consumption |
|
|
|
|
|
|
|
|
| Never
| 4 (7.4%) |
0 (0%) |
4 (13.8%) |
0.1116 |
5 (7.9%) |
3 (8.8%) |
2 (6.9%) |
1.000 |
| Ever |
48 (89.0%) |
25 (100%) |
23 (79.3%) |
56 (88.9%) |
31 (91.2%) |
25 (86.2%) |
|
| Missing |
2 (3.7%) |
- |
2 (6.9%) |
2 (3.2%) |
- |
2 (6.9%) |
|
| Tobacco Consumption |
|
|
|
|
|
|
|
|
| Never |
5 (9.3%) |
2 (8.0%) |
3 (10.3%) |
0.6848 |
6 (9.5%) |
4 (11.8%) |
2 (6.9%) |
0.6848 |
| Ever |
47 (87.0%) |
23 (92.0%) |
24 (82.8%) |
55 (87.3%) |
30 (88.2%) |
25 (86.2%) |
|
| Missing |
2 (3.7%) |
- |
2 (6.9%) |
2 (3.2%) |
- |
2 (6.9%) |
|
| CLINICAL DATA |
|
|
|
|
|
|
|
|
| Tumor Location |
|
|
|
|
|
|
|
|
| Proximal esophagus |
4 (7.4%) |
3 (12.0%) |
1 (3.5%) |
|
5 (7.9) |
4 (11.8%) |
1 (3.4%) |
|
| Middle esophagus |
25 (46.3%) |
11 (44.0%) |
14 (48.3%) |
0.6837 |
26 (41.3%) |
14 (41.2%) |
12 (41.5%) |
0.6211 |
| Distal esophagus |
9 (16.7%) |
4 (16.0%) |
5 (17.2%) |
|
16 (25.4%) |
8 (23.5%) |
8 (27.6%) |
|
| More than one region affected |
12 (22.2%) |
5 (20.0%) |
7 (24.1%) |
|
13 (20.6%) |
6 (17.6%) |
7 (24.1%) |
|
| Missing |
4 (7.4%) |
2 (8.0%) |
2 (6.9%) |
|
3 (4.8%) |
2 (5.9%) |
1 (3.4%) |
|
| Tumor Differentiation |
|
|
|
|
|
|
|
|
| Well and Moderately |
33 (61.0%) |
15 (60.0%) |
18 (62.1%) |
1.000 |
38 (60.3%) |
21 (61.8%) |
17 (58.6%) |
1.000 |
| Poorly and Undifferentiated |
14 (26.0%) |
7 (28.0%) |
7 (24.1%) |
17 (27.0%) |
10 (29.4%) |
7 (24.1%) |
|
| Missing |
7 (13.0%) |
3 (12.0%) |
4 (13.8%) |
8 (12.7%) |
3 (8.8%) |
5 (17.3%) |
|
| Stage |
|
|
|
|
|
|
|
|
| I + III |
18 (33.3%) |
8 (32.0%) |
10 (34.5%) |
1.000 |
19 (30.1%) |
10 (29.4%) |
9 (31.0%) |
1.000 |
| III + IV |
32 (59.3%) |
15 (60.0%) |
17 (58.6%) |
40 (63.5%) |
22 (64.7%) |
18 (62.1%) |
|
| Missing |
4 (7.4%) |
2 (8.0%) |
2 (6.9%) |
|
4 (6.4%) |
2 (5.9%) |
2 (6.9%) |
|
| T stage |
|
|
|
|
|
|
|
|
| T1 + T2 |
5 (9.2%) |
2 (8.0%) |
3 (10.3%) |
1.000 |
5 (7.9%) |
2 (5.9%) |
3 (10.3%) |
0.6586 |
| T3 + T4 |
41 (76.0%) |
20 (80.0%) |
21 (72.4%) |
50 (79.4%) |
27 (79.4%) |
23 (79.4%) |
|
| Missing |
8 (14.8%) |
3 (12.0%) |
5 (17.3%) |
8 (12.7%) |
5 (14.7%) |
3 (10.3%) |
|
| Lymph node invasion |
|
|
|
|
|
|
|
|
| No |
25 (46.3%) |
13 (52.0%) |
12 (41.4%) |
0.5613 |
31 (49.2%) |
18 (52.9%) |
13 (44.8%) |
0.7880 |
| Yes |
22 (40.7%) |
9 (36.0%) |
13 (44.8%) |
|
25 (39.7%) |
13 (38.3%) |
12 (41.4%) |
|
| Missing |
7 (13.0%) |
3 (12.0%) |
4 (13.8%) |
|
3 (8.8%) |
4 (13.8%) |
|
| Overall survival (24 months) |
|
|
|
|
|
|
|
|
| Mean (min-max) |
13 (0.2-99.0) |
7 (0.2-99.0) |
15.0 (0.6-82.8) |
0.0743 |
12.5 (0.2-99) |
7.7 (0.2-75.7) |
15 (0.6-99.0) |
0.2409 |
*calculated with known numbers; # number of patients. Low: expression log2 fold-change < -1; High: expression log2 fold-change > 1.
Supplementary table S3: Association between p21CIP1 mRNA expression and clinical and socio-demographic parameters.
| SOCIO-DEMOGRAPHICDATA |
p21CIP1 |
|
TOTAL (n#=75) |
High (n#=26) |
Low (n#=49) |
p value* |
| Age |
| Median (min-max) |
56.0 (34-83) |
55.0 (34-76) |
56.0 (40-83) |
0.6924 |
| Gender |
|
|
|
|
Men |
61 (81.3%) |
22 (84.6%) |
39 (79.6%) |
0.7590 |
| Women |
14 (18.7%) |
4 (15.4%) |
10 (20.4%) |
| Origin |
|
|
|
|
| Southeast |
46 (61.3%) |
17 (65.4%) |
29 (59.2%) |
0.2628 |
| South |
29 (38.7%) |
9 (34.6%) |
20 (40.8%) |
|
| Alcohol Consumption |
|
|
|
|
| Never |
7 (9.3%) |
1 (3.8%) |
6 (12.2%) |
0.4094 |
| Ever |
66 (88.0%) |
25 (96.2%) |
41 (83.7%) |
| Missing |
2 (2.7%) |
- |
2 (4.1%) |
| Tobacco Consumption |
|
|
|
|
| Never |
8 (10.6%) |
3 (11.5%) |
5 (10.2%) |
1.0000 |
| Ever |
65 (86.7%) |
23 (88.5%) |
42 (85.7%) |
| Missing |
2 (2.7%) |
- |
2 (4.1%) |
| CLINICAL DATA |
|
|
|
|
| Tumor Location |
|
|
|
|
| Proximal esophagus |
5 (6.7%) |
3 (11.5%) |
2 (4.1%) |
|
| Middle esophagus |
33 (44.0%) |
11 (42.4%) |
22 (44.9%) |
0.3243 |
| Distal esophagus |
17 (22.6%) |
3 (11.5%) |
14 (28.6%) |
|
| More than one region affected |
15 (20.0%) |
5 (19.2%) |
10 (20.4%) |
|
| Missing |
5 (6.7%) |
4 (15.4%) |
1 (2.0%) |
|
| Tumor Differentiation |
|
|
|
|
| Well and Moderately |
46 (61.3%) |
16 (61.5%) |
30 (61.2%) |
1.000 |
| Poorly and Undifferentiated |
19 (25.3%) |
7 (27.0%) |
12 (24.5%) |
| Missing |
10 (13.4%)
| 3 (11.5%)
| 7 (14.3%) |
| Stage |
|
|
|
|
| I + II |
20 (26.7%) |
7 (27.0%) |
13 (26.5%) |
1.000 |
| III + IV |
50 (66.7%) |
16 (88.5%) |
34 (69.4%) |
| Missing |
5 (6.6%) |
3 (11.5%) |
2 (4.1%) |
| T stage |
|
|
|
|
| T1 + T2 |
6 (8.0%) |
3 (11.5%) |
3 (6.1%) |
0.3858 |
| T3 + T4 |
61 (81.3%) |
19 (73.1%) |
42 (85.7%) |
| Missing |
8 (10.7%) |
4 (15.4%) |
4 (8.2%) |
| Lymph node invasion |
|
|
|
|
| No |
34 (45.3%) |
11 (42.3%) |
23 (41.4%) |
0.8000 |
| Yes |
33 (44.0%) |
12 (36.0%) |
21 (46.1%) |
|
| Missing |
8 (10.7%) |
5 (12.0) |
3 (11.5%) |