Introduction
Osteosarcoma (OS) is a malignant bone tumor commonly
found in adolescents and young adults. It originates from primitive transformed cells, characterized by aggressive local growth
and high metastases [1]. Although surgery combined with chemotherapy has greatly improved the prognosis of patients with
osteosarcoma, the prognosis of metastatic or recurrent osteosarcoma remains suboptimal [2]. The prognosis is particularly poor
in patients with metastatic osteosarcoma, whose 5-year survival
rate is <30% [3,4]. In spite of several attempts over the past 20+
years using various chemotherapy regimens for osteosarcoma,
survival rates have remained relatively stable, and there hasn’t
been an effective targeted treatment yet [5]. Given this, elucidating the molecular mechanisms of osteosarcoma occurrence,
proliferation, metastasis, and recurrence are of great significance
for developing effective therapeutic strategies and improving prognosis.
In recent years, growing interest has been shown in the pathophysiology and genetics of osteosarcoma, and various genomic studies employing Whole-Genome Sequencing (WGS) and/
or Whole-Exome Sequencing (WES) have been published. Genetic
heterogeneity, multiple chromosomal abnormalities, mutations,
and the most up- and down-regulated genes can all be discovered
by genome analysis [5,6]. Moreover, recent studies have suggested a connection between abnormal alternative splicing regulation and the occurrence or progression of cancer. However, there
is no research on the RBP-AS regulatory network and its possible
functions from the genome-wide level.
In this paper, we examined the immune infiltration-related RAS
and RNA-binding proteins (RBP) regulation network in metastatic
osteosarcoma on a genome-wide scale.
Materials and Methods
Data Collection
Download the published osteosarcoma transcriptome expression data GSE87624 from the GEO database, the data set cell
samples are derived from patients Osteosarcoma tissue, transcriptome data obtained by high-throughput sequencing of 44
osteosarcoma patients and 3 control bone tissues. Based on the
transcriptome data of primary and metastatic osteosarcomas,
differential expression analysis of RBPs and RASEs was carried
out, and the differentially expressed RBP genes and RASEs were
identified. Co-expression analysis of differentially expressed RBP
and RASE was performed to study the RBP-AS regulatory network
in this disease. 44 osteosarcoma patient samples were determined to compare immune cell types and discover differentially expressed RBPs and alternative splicing events in 23 primary and 9
metastatic osteosarcoma tissues. RBP-related genes were collected in the relevant literature [7-10].
Retrieval and process of public data
The public sequence data files were obtained from the Sequence Read Archive (SRA). Using the NCBI SRA Tool fastq-dump,
SRA Run files were converted to fastq format. Using a FASTX-Tool-kit, the raw readings were cleaned of low-quality bases. The clean
readings were then assessed using FastQC.
Reads alignment and differentially expressed gene (DEG) analysis
HISAT2 used clean reads to align to the mouse genome [11].
Ultimately, read count and fragments per kilobase of exon per
million fragments mapped (FPKM) for each gene were evaluated
using uniquely mapped reads. Using FPKM, the expression levels
of the genes were assessed. We choose the DEseq2 software to
gene differential expression analysis. In order to account for the
variation in Library depth, DEseq2 will model the original reads
and utilize the scaling factor. Then, in order to model the read
count, DEseq2 estimates the gene dispersion and decreases these
estimates to get estimates of dispersion that are more accurate.
Finally, DEseq2 matches the model of a negative binomial distribution, and the Wald test or likelihood ratio test is used to evaluate
the hypothesis. The differentially expressed between two or more
samples can be examined using DEseq2. According to fold change
(FC) and false discovery rate (FDR), the results of the study could
well be utilized to assess if the gene is expressed differently. There
are two critical factors: (1) FC: the ratio of the absolute change in
expression; (2) FDR: The following were the significant differential
expression requirements: FDR ≤ 0.05 and FC ≥ 2 or ≤ 0.5.
Alternative splicing analysis
Using the ABL as a pipeline as previously described, the Alternative Splicing Events (ASEs) and regulated alternative splicing
events (RASEs) between the samples were identified and measured [12,13]. In summary, splice junction readings were used by
ABL to detect 10 different forms of ASEs. Using the alternatives
reads and models reads of the samples as raw data, Fisher’s exact
test was chosen to establish statistical significance for sample pair
comparison. We determined the RASE ratio, which is the changed
ratio of alternatively spliced reads and constitutively spliced reads
between comparable samples. The threshold for RASEs detection
was established at the RASE ratio ≥ 0.2 and the p-value ≤ 0.05.
To assess the significance of the ratio alteration of AS events, the
Student’s t-test for repeated comparison was used. Non-intron retention (NIR) RASEs were defined as events that were significant
at a P-value cutoff of 0.05.
Functional enrichment analysis
Using the KOBAS 2.0 server, Gene Ontology (GO) keywords
and KEGG pathways were found to classify DEGs into functional
categories [14]. The enrichment of each term was determined
by using hypergeometric test and the Benjamini-Hochberg FDR
controlling procedure. The study of functional enrichment of the
sets of selected genes also included Reactome pathway profiling.
Immune cell infiltration analysis tool
For the analysis of immune cell infiltration, we used the IOBR
package in the R package, which was published in frontiers in immunology on July 2, 2021 (IF=7.561). ESTIMATE, CIBERSORT, xCell,
TIMER, IPS, MCPcounter, EPIC, and quantTIseq are 8 published
methods for decoding tumor microenvironment (TME) contexture that are combined in IOBR. Additionally, 255 published signature genes set including the tertiary lymphoid structure, tumor
microenvironment, m6A, microsatellite instability exosomes, and
tumor metabolism were gathered by IOBR. Additionally, IOBR employs a variety of methods for data analysis, variables transformation, feature selection, and supports batch survival analysis and even visualization of corresponding result.
Construction of PPI Network
PPI information of all RBP and RASE that were differentially expressed was downloaded after processing by STRING database. To
create the PPI network, we used the Cytoscape software (correlation coefficient ≥ 0.6).
Other statistical analysis
Principal component analysis (PCA) was performed by R package factoextra to show the clustering of samples with the first
two components. The next-generation sequencing data and genomic annotations were visualized in house-script (sogen) after
normalizing the reads by TPM of each gene in the samples. The
clustering based on Euclidean distance was performed by using
the pheatmap package in R. The Student’s t-test was employed to
compare the two groups.
Results
Analysis of gene expression profile of metastatic and primary
osteosarcoma (OS)
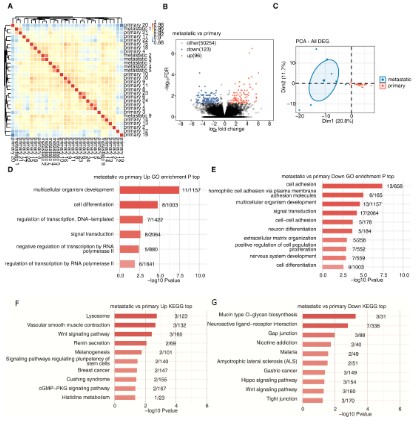
Transcript data of primary osteosarcoma and metastatic osteosarcoma samples were obtained from the dataset GSE87624. A
hierarchical clustering heatmap was used to show a correlation
between metastatic and primary sample based on all expression
genes’ FPKM value (Figure 1A). To find the differentially expressed
genes between primary and metastatic osteosarcoma, we used
differential analysis with the R package limma (FDR ≤ 0.05 and FC≥
2 or ≤ 0.5). The volcano plot showed that 96 genes were up-regulated and 123 genes were down-regulated in metastatic osteosarcoma tissue compared with primary osteosarcoma (Figure 1B).
PCA of all differentially expressed genes in primary and metastatic
osteosarcoma was performed using the R software package factoextra, which clearly showed the results (Figure 1C). For gene
function enrichment analysis, we applied the GO annotations of
the genes in the R package as the background to map the genes
to the background set and enriched them using KOBAS 2.0 server
analysis. The results of the gene set enrichment were then obtained. The GO functional enrichment analysis showed that the up-regulated genes were primarily enriched in the development and
cell differentiation-related pathway of multicellular organisms,
and the down- genes were mainly enriched in the cell adhesion
and extracellular matrix-related pathways (Figure 1D,E).
In addition, the most recent KEGG Pathway gene annotation
was obtained via the KEGG rest API. The same method was adopted to obtain KEGG functional enrichment analysis, which showed
that up-regulated genes were primarily enriched in Lysosomal
and Vascular smooth muscle contraction pathways, while down-regulated genes were mainly enriched in Mucin type O-glycan
biosynthesis and Neuroactive ligand-receptor interaction pathways (Figure 1F,G).
Abnormal alternative splicing patterns in metastatic OS compared with primary OS
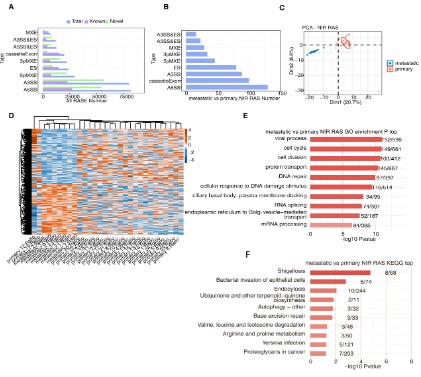
First, we used ABL to calculate the ratio of changes in alternately and constitutively spliced read between the sample, determined as the RASE ratio. RASE ratios ≥0.2 and p-values ≤0.05
were set as thresholds for the detection of RASEs, and significant events with p-values ≤0.05 were considered non-intron-retaining
regulated alternative splicing events (NIR RASEs). On this basis,
we obtained all RASEs in metastatic and primary samples, as
well as 547 NIR RASs that significantly differed between the two
samples (Figure 2A,B). PCA of these NIR RASs was also performed,
clearly distinguishing the two samples (Figure 1C). A hierarchical
clustering heatmap of RAS was drawn based on splicing ratio, and
differences between metastatic and primary osteosarcoma were
clearly identified (Figure 1D). Finally, we used GO and KEGG functional enrichment analysis to enrich the genes corresponding to
these 547 differential RASEs. GO functional enrichment analysis
showed that these genes were mainly enriched in Cell cycle, Cell
division, Protein transport, DNA repair, RNA splicing and other
pathway (Figure 2E). The KEGG functional enrichment analysis indicated that it was mainly enriched in Autophagy, Base excision
repair, Amino acid degradation, and Bacterial infection pathways
(Figure 2F).
Dynamic changes of ASE associated with immune microenvironment regulation in OS
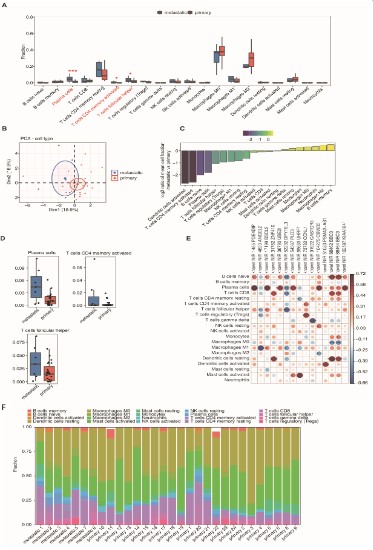
Based on the expression profiles of metastatic and primary
samples, the R software package IOBR package was used to select
the CIBERSORT calculation method and obtain immune infiltrating cell scores for the two groups of samples. Boxplots were used
to show the differences in immune cell types between the two
samples (Figure 3A). Significant increases in T follicular helper cell,
activated memory CD4 T cell, and plasma cell were seen in metastatic osteosarcoma (Figure 3D). This result was slightly different
from the recent results reported by Yang et al. who showed increased numbers of natural killer cells CD56, B cells naive, macrophages M1, and neutrophils in non-metastatic osteosarcoma tissue, while in non-metastatic osteosarcoma tissue macrophages
M2 levels were higher in metastatic tissues [15]. However, this is
consistent with the findings on osteosarcoma lung metastasis reported by Chen et al. [16]. who argued that plasma cells, activated
memory CD4 T cells, T cells CD8, and Tregs were the key determinants of osteosarcoma tissue metastasis. PCA based on fractions
of different immune cells of all expressed genes in the two groups
of samples could also distinguish between the two (Figure 3B). In
addition, the proportion of immune cells in metastatic compared
to primary osteosarcoma showed a decreasing trend (Figure 3C).
The co-expression of RAS and 20 immune cell types was analyzed,
and the pairs with a correlation coefficient ≥ 0.6 were selected.
The regulated alternative splicing genes (RASG) - PDE4DIP, BSCL2,
UBE2I, PLD3, CBWD5, and KIAA1841 were found to be significantly associated with plasma cells. Naive B cells were significantly associated with BSCL2, DPY19L3, BBC3, and KIAA1814. T cells CD8
were significantly associated with ZNF410, DPY19L3, and BBC3.
Both activated CD4 memory T cells and Tregs were significantly
associated with CCNL1. T follicular helper cells were significantly
associated with BSCL2 and DPY19L3 (Figure 3E). Finally, we found
the fractions of different immune cells estimated by Cibersort in
each sample. There were a large number of plasma cells, T cells
CD4, and T cells CD8 in the metastatic osteosarcoma tissue on the
left side of the figure (Figure 3F). In conclusion, we found that after the metastasis of osteosarcoma, the immune infiltrating cells
were mainly divided into plasma cell, CD8 T cell, activated memory CD4 T cell and Tregs.
Differential RBP regulates alternative splicing events related
to immune cells.
In order to obtain differentially expressed RNA-binding protein
(DERBP) genes between metastatic and primary samples, we used
the Venn diagram to intersect 219 DEGs with 2494 RBP genes (obtained from ENCORI database) and finally obtained the top7 DER-BP genes (Figure 4A). Next, we compared the expression of these
DERBP genes in metastatic and primary osteosarcoma by boxplot,
finding that the expressions of CRYAB, FBN1, TRIM61, and RBM20
were significantly down-regulated in metastatic osteosarcoma
tissues, while the expressions of FAM184B and WIPF3 were significantly up-regulated in metastatic osteosarcoma tissues (Figure
4B). In order to understand the relationship between DERBP,
RASE, RASG, and RAS-related immune cells, we used a Network
diagram, which revealed that five DERBPs (WIPF3, FAM184B,
RBM20, TRIM61, CRYAB) were associated with RAS-related immune cells (Figure 4C). The boxplot of the RAS splicing ratio associated with immune cells showed that the splicing ratio of ZNF410
was significantly down-regulated in metastatic osteosarcoma tissues, and the splicing ratio of BSCL2, CBWD5, and KIAA1841 was
significantly up-regulated in metastatic cancer tissues (Figure 4D).
Among them, the alternative splicing event ZNF410 regulated by
CYRAB has negatively correlated with T cells CD4 memory activated, and the alternative splicing event BSCL2 regulated by WIPF3
was positively correlated with plasma cells. Finally, we found that
the read distribution of NIR 17199 BSCL2 and NIR 31762 ZNF410
alternative splicing events were related to immune cells (Figure
4E-F). In conclusion, we speculate that CRYAB and WIPF3 may affect the composition of immune cells by regulating gene alterna-
tive splicing, thereby promoting the metastasis of osteosarcoma
tissue.
RBPs are protein that bind to RNAs through globular RNA-Binding Domains (RBDs), thereby altering the function or fates of the
bound RNAs [17]. RBPs can recognize special RNA-binding domains to interact with RNA and participate in various post-transcriptional regulatory processes, such as RNA splicing, transportation, polyadenylation, intracellular localization, translation, and
degradation [18]. RNA alternative splicing refers to the process
of transcribed precursor mRNA by removing introns and retaining
exons to form mature mRNA, which is a vital step in regulating
post-transcriptional gene expression. As regulation has an affect
on more over 90% of human genes, including genes related with
tumors [19].
The fundamental mechanism of RBP expression and its possible roles are revealed, which helps in the discovery of novel therapeutic targets as well as innovative methods or ideas. DDX24,
DDX21, and IGF2BP2 in RBPs are associated with the prognosis
of osteosarcoma, and WARS may have an important role in the
immune infiltration of osteosarcoma [20]. PUM2 expression was
shown to be low in osteosarcoma patients, and Hu et al. found
that increasing PUM2 expression might inhibit osteosarcoma cells
from migrating and progressing [21]. Moreover, IGF2BP1 expression is up-regulated in osteosarcoma tissues and seems closely
related to the poor prognosis of patients with osteosarcoma
[22]. Pan et al. found that the expression of HuR is significantly
increased in osteosarcoma tissues, and inhibition of HuR could inhibit the viability, EMT, and promote apoptosis of osteosarcoma
cells [23]. A recent study suggested PTBP1 as an oncogene in various cancers [24]. PTBP1 expression was vastly higher in chemotherapy-resistant than chemotherapy-sensitive osteosarcoma tissues, while PTBP1 knockdown enhanced the anti-proliferative and
apoptosis-inducing effects of cisplatin in MG-63 and U2OS. Transcriptome sequencing showed that knockdown of PTBP1 could
up-regulate the expression of copper transporter SLC31A1, and
immunoprecipitation experiments showed that PTBP1 influences
the expression level of SLC31A1 by affecting the stability of the
SLC31A1 mRNA. According to Niu et al. [25], osteosarcoma tissues have higher levels of MSI1 expression than adjacent tissues,
and MSI1 knockdown in osteosarcoma cells can inhibit cancer cell
proliferation and tumor formation. MSI1 was able to bind to the
3'UTR sections of the p21 and p27 mRNAs, according to luciferase
experiments. RBM10 has long been regarded as a tumor suppressor due to its ability to control the MDM2-p53 negative feedback
loop, inhibits the expression of apoptosis proteins like Bcl-2 and
Bax, and promotes the expression of caspase-3 and the production of TNF-α, thereby inducing osteosarcoma cell apoptosis and
inhibits cell proliferation via Notch signaling and the rap1a/Akt/
CREB pathway [26].
Aberrant alternative splicing is prevalent in osteosarcoma,
and its regulation has an important role in the development of
osteosarcoma. There are previous reports of aberrant alternative splicing regulation of some genes. For example, loss or frequent down-regulation of cellular expression of leptin receptor
overlapping transcripts may be associated with tumor formation.
Rothzerg et al., [27] analyzed the AS and transcriptional events
between tumor and normal samples and discovered that up-regulating the expression of IL-6 and TNF-a via overlapping transcription of leptin receptors may influence the occurrence and
metastasis of OS. SRSF3, a member of the Serine/Arginine-Rich
(SR) protein family, regulates gene expression of FoxM1, PLK1,
and CDC25B as well as protein translation, pri-miRNA processing,
polymerization, polyadenylation, and regulates RNA alternative
splicing in U2OS osteosarcoma cells [28]. In human osteosarcoma U2OS cells, Ajiro et al. [29] presented a genomic map of
SRSF3-regulated RSAE and gene expression, whose major transcripts contain highly conserved RNA motifs, revealing that splicing
events were mainly associated with cell proliferation or cell cycle.
Osteosarcoma (OS) is a representative tumor associated with the
Human Telomerase Enzyme Reverse Transcriptase (hTERT) gene,
whose Telomere Maintenance Mechanism (TMM) includes two
forms of Telomerase Activity (TA) and alternative lengthening
telomere (ALT). Hitomi et al. showed that the control of hTERT
expression includes both transcriptional and post-transcriptional processes, both of which contribute to the occurrence of TMM
(TA and ALT) in OS and may provide insight into the prognosis of
patients [30]. In conclusion, both RBPs and AS link the exons of
pre-mRNA in different arrangements, making gene expression
patterns more complex, transcriptionally efficient, and promoting
protein diversity. This eventually results in structurally and functionally distinct mRNA and protein variants and has an important
role in disease.
RASE associated with immune infiltration in OS
With changes in the host immune system, the functional
components of tumor-infiltrating immune cells (TIICs) undergo
minor changes, and TIICs have been reported to be associated
with clinical outcomes in cancer patients [31]. Osteosarcoma
is an immune-sensitive type of tumor, mainly infiltrated by heterogeneous immune cells such as neutrophils, dendritic cells,
monocytes, mast cells, and macrophages [32-34]. Numerous
studies have reported that TIIC subsets such as NK cells, memory
T cells, and M1 macrophages are typically associated with good
prognosis in osteosarcoma, whereas M2 macrophages and Treg
cells are associated with terrible prognosis in osteosarcoma [35-
37]. Additionally, CD4+ memory T cells, CD8+ T cells, NK cells, M1
macrophages, Treg cells, and plasma cells were identified in metastatic tissues as key determinants of osteosarcoma metastasis
[38,39], which is consistent with our results. Chen et al. found
that patrolling mononuclear cells (PMOs) inhibited lung metastasis of osteosarcoma while T follicular helper cell, monocytes, and
resting mast cells were associated with favorable chemotherapy
outcomes for osteosarcoma [16]. In the complex tumor ecology,
in addition to immune cells, there are stromal cell subsets that
can drive malignant tumor progressions, such as endothelial cells,
fibroblasts, reactive astrocytes, and microglia. Peng et al., [40]
explored the potential mechanism through which the predictive
splicing factor affects the overall survival of glioblastoma (GBM)
patients by regulating RASE, and they also found an association
between AS and immune cell infiltration types in tumor tissues
of different subtypes of GBM, establishing that the enrichment of
many immune-related pathways may be caused by differences in
the recruitment or differentiation of various immune cells in malignancies. RAS is closely related to the regulation of the immune
microenvironment during the occurrence of tumors. Therefore,
we studied the correlation between immune cell types and RSAE
in osteosarcoma metastasis at the genome-wide level and analyzed their possible functions.
RASG associated with RBPs in tumors
Lipid droplet morphology is thought to be involved by the endoplasmic reticulum protein, which is encoded by the gene of
BSCL2, and BSCL2 has been linked to both overall survival and
progression-free survival in high-grade ovarian serous carcinoma
(HGOSC) [41]. Ali et al. [42] analyzed ovarian cancer data from
TCGA and showed that in univariate and multivariate analysis,
the expression profile of the gene BSCL2 had a statistically significant correlation with the survival rate of ovarian cancer patients.
However, as the gene has been studied to a lesser extent in osteosarcoma, the specific mechanism remains unknown. A transcription factor (TF) called ZNF410, also referred to as APA-1, regulates
the expression of genes involved in matrix remodeling during the
senescence of fibroblasts [43]. The research on ZNF410 is lacking, yet, the latest cancer research reported the association of abnormal expression of this gene in breast cancer with tumor stage and
different subtypes [44]. CBWD5, also known as CBWD3, is currently only reported to have copy number variation in CBWD5 in small
cell lung cancer [45].
The blank of RBM10/20 and WIPE3 in osteosarcoma research
The expression of RBM10 can induce the apoptosis of osteosarcoma and inhibit the proliferation of primary chondrocytes by
reducing the production of Bcl-2, increasing the production of
caspase-3 and the expression TNF-α. However, over-expression of
Bcl-2 can inhibit osteosarcoma invasion and migration as well as
decrease osteosarcoma colony formation and proliferation [46].
As one of the few heart-specific splicing factors, previous studies
of RBM20, which belongs to the same class, have mostly concentrated on research in cardiomyopathy. Specific genes involved in
sarcomere assembly, ion transport, and relaxation function have
been shown to be regulated by RBM20. It acts on actin and tropomyosin in familial cardiomyopathy, affecting striated muscle
biomechanics. In addition, RBM20 has been implicated in fasting
blood glucose regulation of insulin damage in cardiac tissue [47].
However, the role of RBM20 in osteosarcoma has not been proven so far. WIPE3 is also rarely reported in osteosarcoma and is
currently only reported in a few cancers such as gastric cancer
and breast cancer. Cava et al. [48] argued that with the increased
aggressiveness of breast cancer molecular subtypes, the interaction between DERBPs and DEGs is one of the essential factors for
the future progress of breast cancer research. By analyzing the
microarray data of gastric cancer tissue, suggesting that the abnormal expression of WIPF3 is connected to the survival rate of
patients with gastric cancer, Zhou et al. [49] discovered four RBPs
(RBPMS2, DAZ1, WIPF3, and NOVA1) which independently predicted the prognosis of gastric cancer. However, DAZ1 and WIPF3
have not yet been reported in osteosarcoma, which indicates that
they might be potential therapeutic targets and prognostic indicators for osteosarcoma. We found that alternative splicing events
regulated by WIPF3-BSCL2 were positively associated with plasma
cells in metastatic osteosarcoma tissue. Nonetheless, it remains
unclear how WIPF3 affects splicing complex formation and pre-mRNA structure after binding to target gene sequences. In the
future, we plan to conduct a more in-depth study on the specific
molecular mechanism of WIPF3 regulating the alternative splicing
of target genes.
Conclusion
We identified a total of 547 differentially alternative splicing
events in metastatic osteosarcoma tissues, screened the top 7
DERBPs and associated alternative splicing events with different
types of immune cells. Finally, analyzed their co-expression relationships. Among them, the alternative splicing event ZNF410
regulated by CYRAB has negatively correlated with T cells CD4
memory activated, and the alternative splicing event BSCL2 regulated by WIPF3 was positively correlated with plasma cells. In
conclusion, we speculate that CRYAB and WIPF3 may affect the
composition of immune cells by regulating gene alternative splicing, thereby promoting the metastasis of osteosarcoma tissue.
Does ZNF410 or WIPF3 affect the formation of splicing complexes
after binding to target genes to regulate the alternative splicing
process? And the specific molecular mechanism of ZNF410 or WIPF3 regulating target gene alternative splicing needs further
study.
Declarations
Conflicts of interest: The authors declare that the research
was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability statement: Publicly available datasets were
analyzed in this study. This data can be found here: The data for
this study can be found in the GEO database (https://www.ncbi.
nlm.nih.gov/geo/query/acc.cgi?acc=GSE87624).
Funding Statement: The National Natural Science Foundation
of China (81401790).
References
- Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and
progression of childhood solid tumours. Mol Cancer. 2017; 16: 15.
- Chen C, Xie L, Ren T, Huang Y, Xu J, et al. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent
breakthroughs. Cancer Lett. 2021; 500: 1-10.
- Lv T, Jian Z, Li D, Ao R, Zhang X, et al. Oxyresveratrol induces apoptosis and inhibits cell viability via inhibition of the STAT3 signaling
pathway in Saos-2 cells. Mol Med Rep. 2020; 22: 5191-5198.
- Liao S, Zhou S, Wang C. GAPLINC is a predictor of poor prognosis
and regulates cell migration and invasion in osteosarcoma. Biosci
Rep. 2018; 38.
- Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, et al.
Molecular Biology of Osteosarcoma. Cancers (Basel). 2020; 12.
- Ho XD, Phung P, V Q. Le, V H. Nguyen, Reimann E, Prans E, Kõks G,
et al. Whole transcriptome analysis identifies differentially regulated networks between osteosarcoma and normal bone samples.
Exp Biol Med (Maywood). 2017; 242: 1802-1811.
- Castello A, Fischer B, Eichelbaum K. Insights into RNA biology from
an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:
1393-1406.
- Castello A, Fischer B, Frese Ck. Comprehensive Identification of
RNA-Binding Domains in Human Cells. Mol Cell. 2016; 63: 696-
710.
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding
proteins. Nat Rev Genet. 2014; 15: 829-845.
- Hentze M W, Castello A, Schwarzl T. A brave new world of RNA-
binding proteins. Nat Rev Mol Cell Biol. 2018; 19: 327-341.
- Kim D, Pertea G, Trapnell C. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14: R36.
- Jin L, Li G, Yu D. Transcriptome analysis reveals the complexity of
alternative splicing regulation in the fungus Verticillium dahliae.
BMC Genomics. 2017; 18: 130.
- Xia H, Chen D, Wu Q. CELF1 preferentially binds to exon-intron
boundary and regulates alternative splicing in HeLa cells. Biochim
Biophys Acta Gene Regul Mech. 2017; 1860: 911-921.
- Xie C, Mao X, Huang J. KOBAS 2.0: a web server for annotation and
identification of enriched pathways and diseases. Nucleic Acids
Res. 2011; 39: W316-W322.
- Yang B, Su Z, Chen G. Identification of prognostic biomarkers associated with metastasis and immune infiltration in osteosarcoma.
Oncol Lett. 2021; 21: 180.
- Chen T, Zhao L. Patrolling monocytes inhibit osteosarcoma metastasis to the lung. Aging (Albany NY). 2020; 12: 23004-23016.
- Montalbano M, Mcallen S, Sengupta U. Tau oligomers mediate aggregation of RNA-binding proteins Musashi1 and Musashi2 inducing Lamin alteration. Aging Cell. 2019; 18: e13035.
- Glisovic T, Bachorik J L, Yong J. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008; 582: 1977-1986.
- Coomer A O, Black F, Greystoke A. Alternative splicing in lung cancer. Biochim Biophys Acta Gene Regul Mech. 2019; 186: 194388.
- Li B, Fang L, Wang B. Identification of Prognostic RBPs in Osteosarcoma. Technol Cancer Res Treat. 2021; 20: 2091220490.
- Hu R, Zhu X, Chen C. RNA-binding protein PUM2 suppresses osteosarcoma progression via partly and competitively binding to
STARD13 3’UTR with miRNAs. Cell Prolif. 2018; 51: e12508.
- Wang L, Aireti A, Aihaiti A. Expression of microRNA-150 and its
Target Gene IGF2BP1 in Human Osteosarcoma and their Clinical
Implications. Pathol Oncol Res, 2019; 25: 527-533.
- Pan W, Pang J, Ji B. RNA binding protein HuR promotes osteosarcoma cell progression via suppressing the miR-142-3p/HMGA1 axis.
Oncol Lett. 2018,16: 1475-1482.
- Cheng C, Ding Q, Zhang Z. PTBP1 modulates osteosarcoma chemoresistance to cisplatin by regulating the expression of the copper transporter SLC31A1. J Cell Mol Med. 2020; 24: 5274-5289.
- Niu J, Zhao X, Liu Q. Knockdown of MSI1 inhibited the cell proliferation of human osteosarcoma cells by targeting p21 and p27.
Oncol Lett. 2017; 14: 5271-5278.
- Cao Y, Di X, Zhang Q. RBM10 Regulates Tumor Apoptosis, Proliferation, and Metastasis. Front Oncol. 2021; 11: 603932.
- Rothzerg E, Ho X D, Xu J. Alternative splicing of leptin receptor
overlapping transcript in osteosarcoma. Exp Biol Med (Maywood).
2020; 245: 1437-1443.
- Jia R, Li C, Mccoy Jp. SRp20 is a proto-oncogene critical for cell
proliferation and tumor induction and maintenance. Int J Biol Sci.
2010; 6: 806-826.
- Ajiro M, Jia R, Yang Y. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS
cells[J]. Nucleic Acids Res. 2016; 44: 1854-1870.
- Fujiwara-Akita H, Maesawa C, Honda T. Expression of human telomerase reverse transcriptase splice variants is well correlated
with low telomerase activity in osteosarcoma cell lines. Int J Oncol.
2005; 26: 1009-1016.
- Shibutani M, Maeda K, Nagahara H. Tumor-infiltrating Lymphocytes Predict the Chemotherapeutic Outcomes in Patients with
Stage IV Colorectal Cancer. In Vivo. 2018; 32: 151-158.
- De PALMA M, LEWIS C E. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013; 23: 277-286.
- Mantovani A, Marchesi F, Malesci A. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;
14: 399-416.
- Kelleher F C, O’sullivan H. Monocytes, Macrophages, and Osteoclasts in Osteosarcoma. J Adolesc Young Adult Oncol. 2017; 6: 396-
405.
- Rosenberg J, Huang J. CD8(+) T Cells and NK Cells: Parallel and
Complementary Soldiers of Immunotherapy. Curr Opin Chem Eng.
2018; 19: 9-20.
- Sica A, Larghi P, Mancino A. Macrophage polarization in tumour
progression. Semin Cancer Biol. 2008; 18: 349-355.
- Saito T, Nishikawa H, Wada H. Two FOXP3(+) CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat
Med. 2016; 22: 679-684.
- Gao K, Li X, Zhang L. Transgenic expression of IL-33 activates CD8(+)
T cells and NK cells and inhibits tumor growth and metastasis in
mice[J]. Cancer Lett. 2013; 335: 463-471.
- Zhang Y L, Li Q, Yang XM. SPON2 Promotes M1-like Macrophage
Recruitment and Inhibits Hepatocellular Carcinoma Metastasis by
Distinct Integrin-Rho GTPase-Hippo Pathways. Cancer Res. 2018;
78: 2305-2317.
- Zhao L, Zhang J, Liu Z. Comprehensive Characterization of Alternative mRNA Splicing Events in Glioblastoma: Implications for
Prognosis, Molecular Subtypes, and Immune Microenvironment
Remodeling. Front Oncol. 2020; 10: 555632.
- Cuello MA, Kato S, Liberona F. The impact on high-grade serous
ovarian cancer of obesity and lipid metabolism-related gene expression patterns: the underestimated driving force affecting prognosis. J Cell Mol Med. 2018; 22: 1805-1815.
- Hossain M A, Saiful I S, Quinn J. Machine learning and bioinformatics models to identify gene expression patterns of ovarian cancer
associated with disease progression and mortality. J Biomed Inform. 2019; 100: 103313.
- Benanti J A, Williams D K, Robinson K L. Induction of extracellular
matrix-remodeling genes by the senescence-associated protein
APA-1. Mol Cell Biol. 2002; 22: 7385-7397.
- Li X, Zhu J, Qiu J. Identification of Potential Prognostic Biomarkers
for Breast Cancer Based on lncRNA-TF-Associated ceRNA Network
and Functional Module. Biomed Res Int. 2020; 2020: 5257896.
- Zhou H, Hu Y, Luo R. Multi-region exome sequencing reveals the
intratumoral heterogeneity of surgically resected small cell lung
cancer. Nat Commun. 2021; 12: 5431.
- Cao Y, Di X, Zhang Q. RBM10 Regulates Tumor Apoptosis, Proliferation, and Metastasis. Front Oncol. 2021; 11: 603932.
- Guo W, Schafer S, Greaser ML. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012; 18: 766-773.
- Cava C, Armaos A, Lang B. Identification of long non-coding RNAs
and RNA binding proteins in breast cancer subtypes. Sci Rep. 2022;
12: 693.
- Zhou L, Zhou Q, Wu Y. Integrating 13 Microarrays to Construct a 6
RNA-binding proteins Prognostic Signature for Gastric Cancer patients. J Cancer. 2021; 12: 4971-4984.