Introduction
Colorectal Cancer (CRC), the most common malignant tumor
of the digestive tract, has expanded remarkably swiftly in recent
years and is linked to high rates of morbidity and mortality as well
as ease of recurrence and metastasis [1]. Per the CRC treatment
guidelines, surgery and adjuvant chemotherapy are the primary
treatment options. Particularly for patients with terminal CRC,
surgical therapy for CRC has a risk of recurrence and metastasis,
as well as surgical aftereffects including diarrhea or constipation.
Cisplatin (DDP) is a potent chemotherapy drug that has a wide
anti-cancer spectrum and is not cell-cycle dependent. It is a
component of a platinum-based metal complex that inhibits
the transcription and replication of intracellular DNA or binds
to nuclear proteins and plasma proteins to have an anti-cancer action [2,3]. However, its widespread use is restricted due to its
toxicity to healthy cells and other harmful side effects. Therefore,
the development of anti-tumor medicines with novel modes of
action and low toxicity has garnered interest to suppress the
development of CRC and prevent tumor spread and recurrence.
Traditional Chinese medicine is a cultural gem of China.
More academics are focusing on the clinical research of natural
medicine in the area of cancer because of the benefits of TCM
treatment based on syndrome distinction, treatment of non-diseases, and multi-target therapy. The mechanism of action of
TCM is unclear due to its complex chemical components, multiple
targets, and multi-level pharmacological effects. However, a novel
tool and approach for determining the mechanism of therapeutic
action are provided, which stimulates the modernization and
globalization of TCM. It combines the target database of TCM in
many diseases with molecular docking verification and network
pharmacology. The development and application of systems
biology, which includes contemporary genomics, proteomics,
and metabolomics, is where the idea of network pharmacology
originated. To support and offer a significant amount of data
on the interaction of biological entities, large-scale biological
databases are required. The traditional search for a single target
in pharmacological research will give way to thorough network
analysis, such as through the creation of a «target drug-disease-gene» network. A new tool and concept for analyzing the
mechanism of drug action were developed by analyzing and
observing the influence of drugs on diseases [4,5]. This method is
similar to «treating the same disease with different diseases» and
«treating the same disease with the same disease» in traditional
Chinese medicine theory.
Artemisia annua was included as a remedy for malaria in Ge
Hong’s «handbook of medicines for emergencies» during the
Eastern Jin Dynasty: «Artemisia annua a grip, with two liters of
water stains, wring juice, to drink it.» Artemisinin is the most
effective medication for treating malaria resistance. Artemisinins
and their derivatives display powerful anticancer potential in
addition to their anti-malarial properties, according to more
recent studies [6]. DHA is one of the most studied derivatives
used to treat malignancies. Studies have demonstrated that
DHA has inhibitory effects on the proliferation and growth of a
range of tumor cells, including cervical cancer cells, lung cancer
cells, oesophageal squamous carcinoma cells, gastric cancer
cells, and liver cancer cells [7-11], supporting the idea that DHA
is a potentially effective anti-tumor agent. However, few in vivo
research on DHA in the therapy of CRC have been described, and
the majority of these are in vitro.
In this study, the inhibitory impact of DHA on subcutaneously
transplanted tumors in nude mice was assessed. The inhibitory
impact of DHA on CRC cells and its probable mechanism were
then evaluated using network pharmacology and molecular
docking approaches. Using the RKO cell line, we checked into
whether DHA may trigger endogenous apoptosis and block the
cell cycle in the G2/M phase by inhibiting the PI3K/AKT signaling
pathway. These results will serve as an experimental foundation
for the application of DHA in the therapy of CRC.
Materials and methods
Drugs and reagents
DHA was synthesized and gifted by the group of Zhang Wanbin
of Shanghai Jiaotong University with a purity of >99%. 25 mg
of Cisplatin/stem (Sigma-Aldrich, USA). A 0.2 mg/ml solution
was mixed with ordinary saline before use. The solution was
ultrasonically solubilized and stored at 4°C.
Animals
Sixty male nude rats of SPF grade, 6-7 weeks old, weighing 24 ±
2 g, were purchased from Shanghai Siple-Bikai Laboratory Animal
Co.
All experimental protocols involved were approved by the
ethics committee of the Shanghai University of Traditional Chinese
Medicine (PZSHOTCM201113011). The experimental animals
were kept under standard housing conditions, with a temperature
of 20-25°C, Relative humidity was maintained between 40% and
60%, and normal day and night cycles.
Tumor xenograft model
RKO human colorectal cancer cells were altered to 1 × 107
cells/ml. To establish a subcutaneous xenograft tumor model
in nude mice, the mixed cells were resuspended and injected
subcutaneously into the left axilla of the animals using 0.2 ml of cell
suspension. In nude mice, the development of xenograft tumors
was observed and recorded. Mice were randomly allocated to
one of four groups after model building. Drug delivery was begun
after the tumor’s diameter reached about 5 mm.
Three times per week, a 2 mg/kg intraperitoneal injection of
cisplatin was given to the Cisplatin group. The DHA groups received
either a high dose (200 mg/kg/d) or a low dose (100 mg/kg/d) of
DHA. The model group received 10 ml/kg/d of normal saline. The
medication was given constantly for 30 days. Daily observations
of mice’s activity, mental state, skin, and diet were made by
modeling. The medication was given when the transplanted
tumor’s diameter tumor was more than 5 mm. The major diameter
(L) and transverse diameter (W) of the transplanted tumor were
measured and recorded with electronic vernier calipers. The
tumor volume was calculated using the formula: tumor volume
(V,mm3)=[l(mm) X W2(mm2)]/2. The tumors’ growth curve was
depicted. the pharmacological intervention was terminated on
the 31st day, and the blood was collected from the intraocular
venous plexus of the mice. The mice were sacrificed. The mice’s
subcutaneous transplanted tumors underwent exfoliation and
weighing. Utilizing the formula, the tumor inhibition rates of
different groups were obtained:
tumor inhibition rate (%) = (1 − Treatment Group average
tumor weight/Model Group average tumor weight) ×100%.
Hematoxylin-eosin staining (H&E)
The exfoliated tumors were stored at 4°C after being fixed in
a 4% paraformaldehyde solution. The microtome rack held the
prepared tumor paraffin slices, which were then moved to the
staining box. The staining steps are as follows: xylene, 10 min
twice; absolute ethanol, 5 min twice; 95% ethanol, 5 min; 90%
ethyl alcohol, 5 min; 80% ethanol, 5 min; 70% ethanol, 5 min; Washed repeatedly with distilled water and drain. The sections
were stained with hematoxylin for 3-8 min and then washed under
running water. Differentiation with 1% HCL for a brief period of
time, followed by a rinse under running water; 0.6% ammonia
water converted the rinse back to blue. Eosin dye was used to
stain the sections for 1-3 min, after which 95% ethanol was added
successively for 5 min, twice. Absolute ethanol, twice for 5 min
each, as well as xylene. Following drying, the portions were
dehydrated and coated with neutral gum. Images were gathered
for examination after being examined under a microscope.
Enzyme-linked immunosorbent assay for TNF-α levels
Before the execution of each group of mice, blood was drawn
from the posterior ocular plexus into 1.5 ml EP tubes in an amount
of about 0.5 ml. Each tube containing a sample of whole blood
had its information marked. They were kept there overnight at
4oC. The blood was centrifuged at 1000 rpm for 15 minutes to
prevent repeated freezing and thawing. The supernatant was
then separated into new EP containers and kept at -80°C. Thawed
samples should be centrifuged before testing them. According
to the instructions on the kit, serum samples from each group
should be taken to evaluate the TNF-α (NeoBioscience, China).
Target prediction of DHA and network construction
The CAS number of DHA (71939-50-9) was input into the
PubChem [12] (https:// pubchem.ncbi.nlm.nih.gov) for retrieval
of DHA, and the SMILES number [CC1CCC2C(C(OC3C24C1CCC(O3)
(OO4)C)O)C] and structural data were saved. SMILES number of
DHA was input into SWISS [13] (http://www.swisstargetprediction.
ch/index.php), «Homo sapiens» was selected to limit the source of
screened species, and target prediction was carried out based on
structural similarity. The target in prediction results was selected
and set as DHA target file.csv. Select «Ingredient» in SymMap [14]
(https://www.symmap.org/) and enter « dihydroartemisinin» to
retrieve. Then the targets obtained from the two databases were
summarized and de-processed to integrate a comprehensive drug
target database. «Colon cancer» was entered into GeneCards
[15] (https://www. genecards.org) for retrieval, and the obtained
disease gene target score was sorted in descending order.
The obtained DHA predicted target genes and colon cancer
gene targets were respectively input into the corresponding
positions in the Draw Venn Diagram (http://bioinformatics.
psb.ugent.be/webtools/Venn/), and the gene intersection was
compared, and the Venn diagram was drawn, and the common
cross-target genes were extracted as potential targets of DHA in
the treatment of CRC.
In order to collect the PPI Network data, the common target
genes following the intersection were submitted to String
(https://string-db.org/cgi/input.pl) [16]. The PPI network data
was then used to construct the drug-disease target protein
interaction network using Cytoscape software. The Network
Analyzer function of the Degree value was used to evaluate the
PPI network and modify the size and color of each target node in
the PPI network (connection degree).
Molecular docking analysis
Prepare protein Using the «Prepare Protein» function in the
Discovery Studio 4.0 software, the active sites were defined after the protein was dehydrated, hydrogenated, repaired the defect
area, and optimized the structure. Since the protein crystal
structure used for molecular docking this time, all contain small
molecule inhibitors, the binding site of each small molecule
inhibitor is directly defined as the active site, and the radius of
the active site is set to 5-10 A according to different situations of
each protein.
Small molecule preparation Using the «prepare ligands»
function in the software, the small molecule was optimized, and
conformation search was conducted, and only one conformation
was obtained when the DHA molecule was prepared. To prepare
for molecular docking, the conformation with the minimum
energy was determined after «full minimization» had optimized
the conformation’s energy. The software’s «CDOCKER» function
was used to perform molecular docking, with RMSD defined to
0.5 and the other parameters left to their default values.
Enrichment analysis
To undertake the GO biological process and KEGG pathway
enrichment analysis of the potential target of DHA in the CRC
treatment, the common target genes were imported into the
David [17] (https://david.ncifcrf.gov) database, and the species
«Homo sapiens» was chosen. GO is a biological functional system,
which is used to define the function and features of gene products.
For the GO analysis, three modules - biological process (BP), cell
composition (CC), and Molecular Function (MF) - were chosen.
The pathway analysis method of choice was the KEGG module.
KEGG pathway enrichment analysis was created to obtain much
richer biological pathways, more efficiently connect link gene
lists with higher-order functional information, and systematically
examine gene function. By screening the FDR less than 0.05 GO
biological process and KEGG enrichment data (P<0.05), the top 20
signaling pathways were identified. The amount of genes (count
value) enriched in each pathway were ranked in descending
order. Bioinformatics (http://www.bioinformatics.com.cn/) then
displayed the analysis results.
Cell culture
Human colon cancer RKO cells, purchased from the Cell
Resource Center of Shanghai Institutes for Biological Sciences,
Chinese Academy of Sciences. The RKO cell line was cultured in
RPMI 1640 medium containing 10% fetal bovine serum and 1%
penicillin-streptomycin test solution which was placed in a 37oC,
5%_CO2 cell culture incubator. The cells were changed every 1-2 d
according to their growth status and passaged once every 2-3 d.
Cell proliferation assay
RKO cells were planted in 96-well plates at a density of 1 ×
104 cells/ml and 100 μl per well during the logarithmic growth
phase. Based on the concentration of DHA interventions were
administered (0, 25, 50, and 100 μmol/L) (the solution had a
volume of 100 μl, and the 0 μmol/L groups were given a medium
containing equal levels of DMSO). Each well received 20 μl MTT
reagent (5 mg/ml, (Sigma-Aldrich, USA)) after 12, 24, 48, and 72
h of incubation. Incubation was then continued for 4 h. Carefully
removing the culture medium, 150 μl of DMSO was applied to each well. After shaking in the dark for 10 min, the absorbance
(OD) was evaluated at 570 nm in a microplate reader. The
inhibition ratio of cell proliferation was calculated.
Cell apoptosis detection by flow cytometry
At the point of logarithmic growth RKO cells were collected.
After optimizing the cell density to 2.5 × 105 cells/ml and
inoculating the cells at 2 ml/well onto 6-well plates (confocal
dishes), the medium was removed once the cells had become
adherent. Different concentrations (0, 50, 95, 150 μmol/L) of DHA
were given and cells were collected for subsequent detection
after 48 h of incubation. After 48 h of cell grouping and drug
intervention, cells were harvested and washed twice with pre-cooled DPBS. The cells were resuspended by adding 150 μl of
1×Binding Buffer to the cell precipitate to make the cell density
reach 1 × 106 cells /ml. A fresh centrifuge tube was filled with
100 μl of cell suspension, and then 5 l Annexin V-FITC and 10
μl PI reagent (Meilunbio, China) were gently added, combined,
and incubated for 15 min at room temperature in the dark. Flow
cytometry was used to assess cell apoptosis.
Hoechst 33258 staining for detection of cell apoptosis
After 48 h of cell grouping and pharmacological intervention
following «2.11», the growth medium was aspirated, 0.5 ml of
fixation solution was applied, and the cells were fixed overnight at
4°C. The next day, the fixative was discarded after being cleaned
twice for 3 min each with DPBS. 0.5 ml of Hoechst 33258 staining
solution (Meilunbio, China) was applied to stain for 5 min. Then,
it was rinsed twice for 3 min each with 0.5 ml of DPBS. The liquid
was aspirated. The anti-fluorescence quenching solution was
included. A fluorescence microscope was used to capture images
of the cell apoptosis.
Cell cycle assay
After 48 h of cell grouping and medication intervention
following «2.11», cells were harvested. The mixture was
centrifuged at 1,000 rpm for 5 min after adding 1 ml of pre-cooled
DPBS. The supernatant was removed. 1 ml of DPBS was given and
resuspended. 3 ml of pre-cooled anhydrous ethanol was gradually
added dropwise until the ethanol concentration reached 75%,
and stored overnight at 4°C.
According to the kit’s instructions, the staining solution (staining
buffer: PI staining solution: 50×RNase A=100:5:2) (Meilunbio,
China) was prepared in advance and kept at 4°C for later use.
After removing the fixed cell suspension and centrifuging the
supernatant, the cells were resuspended by adding 1 ml of chilled
DPBS.
Each tube of the sample received 500 μl of pre-prepared
staining working solution, and then the cells were gently mixed
before being incubated for 30 min at 37°C in the dark. The
supernatant was then absorbed and discarded. The cell cycle was
determined by flow cytometry.
Wound healing assay
RKO cells were adjusted density to 5 × 106 cells /ml and then
planted in 6-well plates. A 200 μl micropipette was used to create
three parallel wounds perpendicularly to the plate plane after
the cells had become adherent and overgrown. To get rid of the cell debris, DPBS was applied three times to the cells. DHA
was supplied at 0 and 95 μmol/L amounts, respectively. under a
microscope, the wounds were captured at 0, 6, 12, and 24 h. The
area of the blank area was measured by using ImageJ software
and the cell migration rate was calculated.
Real-time quantitative PCR assay
RKO cells at the logarithmic growth stage were obtained, and
the cell density was regulated to 3 × 105 cells/ml. 2 ml/well cells
were then seeded in 6-well plates. DHA at concentrations of 0
and 95 μmol/L was added, respectively. After 12 h of cell grouping
and drug intervention, cells in each group were collected, and
total RNA was extracted by adding 1 ml TRIzol reagent and 200 μl
chloroform. Then reverse transcription reaction was carried out
according to the kit (CWBio, China) instructions to prepare cDNA.
The mRNA expression levels of Caspase-3, Caspase-9, Bcl-2, and
Bax in each group were detected by fluorescence quantitative
PCR with β-actin as the internal reference. All the primers were
synthesized by Shanghai Shenggong Bioengineering Co., LTD. The
sequence of primers is shown in Table 1.
PCR reaction conditions: 95°C for 30 s; 45 cycles of 95°C for 5
s and 60°C for 30 s. Dissolution curve analysis: 95°C, 15 s; 60°C, 1
min; 95°C, 15 s; 50°C for 30 s. The relative mRNA expression level
was quantitatively analyzed by the 2-∆∆CT method.
Table 1: Primer sequences of target genes.
| Primer name |
Specific sequence |
| Caspase-3 |
Forward: 5'-CCAAAGATCATACATGGAAGCG-3' |
| Reverse: 5'-CTGAATGTTTCCCTGAGGTTTG-3' |
| Caspase-9 |
Forward: 5'-GACCAGAGATTCGCAAACCAGAGG-3' |
| Reverse: 5'-AAGAGCACCGACATCACCAAATCC-3' |
| Bax |
Forward: 5'-CGAACTGGACAGTAACATGGAG-3' |
| Reverse: 5'-CAGTTTGCTGGCAAAGTAGAAA-3' |
| Bcl-2 |
Forward: 5'-CATGCTGGGGCCGTACAG-3' |
| Reverse: 5'-GAACCGGCACCTGCACAC-3' |
| β-actin |
Forward: 5'-GGGACCTGACTGACTACCTC-3' |
| Reverse: 5'-TCATACTCCTGCTTGCTGAT-3' |
Western blot
Inoculating cells onto 60 mm culture dishes after adjusting the
cell density to 1 × 107 cells/ml. DHA was added in various concentrations: 0, 50, and 95 μmol/L. After 48 h of drug intervention,
200 μl of mixed lysis solution (RIPA lysis solution with protease
and phosphatase inhibitor) was added, and it was then lysed on
ice for 30 min. The total protein concentration of cells was calculated using the BCA method. polyacrylamide gel electrophoresis
(70 V, 50 min; 120 V, 1 h) was applied to separate each well which
contained 40 μg of protein. Transferring the isolated proteins to
PVDF membranes (0.2 A, 2 h).
5% skim milk was added and blocked on a table concentrator
for 2 h at room temperature. The membrane was cleaned three
times with TBST for 10 min each time; primary antibodies to each
target protein were added (1:1,000 volume dilution) and incubated overnight at 4°C, antibodies to MMP-9 (Batch No. 13667),
Caspase-3 (Batch No. 14220), caspase-9 (Batch No. 9508), cleaved Caspase-3 (Batch No. 9661), cleaved Caspase-9 (Batch No.
7237), Bcl-2 (Batch No. 5023), Bax (Batch No. 4223), PI3K (Batch
No. 4257), AKT (Batch No. 4691), p-PI3K (Batch No. 4228), p-AKT
(Batch No. 4060) and, p38 MAPK (Batch No. 8690), p-P38 MAPK
(Batch No. 4511), and β-actin (Batch No. 3700) all from Cell Signaling were used. The membranes were washed 3 times with
TBST for 10 min each time. Then, primary antibodies were added
(1:1,000 v/v) and reacted overnight at 4°C. The membrane was
cleaned with TBST 3 times the following day for 10 min each time.
Horseradish peroxidase-labeled secondary antibody (volume dilution ratio of 1:5000) was then given and processed for 2 h at room
temperature on a table concentrator. The membrane was then
given 3 TBST washes, each lasting 10 min. Exposure development
was performed with a gel imaging system following the ECL kit’s
instructions. By utilizing the ImageJ image analysis management
system, the protein bands were quantified.
Statistical analysis
SPSS 16.0 software was used for statistical data analysis, and
GraphPad 9.0 software was used for plotting. The measurement
data were expressed as x±SD. The one-way ANOVA was used for
comparison between multiple groups. A T-test was used for multiple comparisons. The difference was considered statistically significant at p<0.05.
Results
Effects of DHA on the growth of RKO tumor-bearing nude
mice
A subcutaneous tumor developed about 9 days after RKO cells
were injected, and the transplanted tumor had a diameter of 5
mm. In comparison to the model group, the mice receiving cisplatin were depressed, sluggish to react, and consumed less food
after delivery. Both the high- and low-dose DHA groups of mice
displayed improved mental health, increased activity, and normal
eating habits. Mice in the cisplatin group demonstrated a significant loss of body mass when compared to the model group after
30 days of the administration (P<0.01), while mice in the low-dose
group showed an increase in body weight (P<0.05); Mice in the
high and low dose DHA groups displayed a considerable increase
in body mass, in contrast to mice in the cisplatin group (P<0.001)
(Table 2). These results indicated that, in contrast to cisplatin, DHA
could greatly improve the tumor-bearing mice's growth status.
Table 2: Effects of DHA on body weight in RKO tumor-bearing
nude mice.
| Group |
Body Weight (g) |
| Before treatment |
After treatment |
| Model |
23.67 ± 1.32 |
30.08 ± 1.52 |
| Cisplatin |
23.63 ± 1.05 |
26.61 ± 1.55** |
| DHA Low Dose |
24.82 ± 0.69 |
32.08 ± 1.33* ### |
| DHA High Dose |
24.81 ± 1.18 |
31.17 ± 2.17### |
(n=10, data represent mean ± SD; **P<0.01, *P<0.05 vs Model; ###P<0.001
vs Cisplatin).
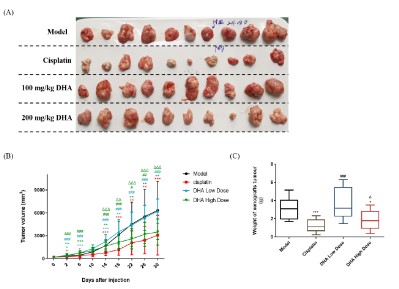
Effects of DHA on the growth of RKO transplanted tumors
The treatment was started when the mice had subcutaneous
transplanted tumors more than 5 mm in diameter. The tumor
volume in each group was measured by vernier calipers every 4
days, and the growth curve of the RKO tumor was displayed in
Figure 1 (A) and (B). The mice were sacrificed on day 31 and the
weight of the exfoliated tumor tissue was recorded. As indicated
in Figure 1 (C), the tumor inhibition rate was evaluated. After administration, the high-dose DHA group and the cisplatin group
significantly inhibited tumor volume as compared to the model
group. The cisplatin group showed the highest rate of tumor inhibition (60.80%), followed by the 200 mg/kg DHA group (41.45%),
while the 100 mg/kg DHA group had no inhibitory impact on the
RKO implanted tumor.
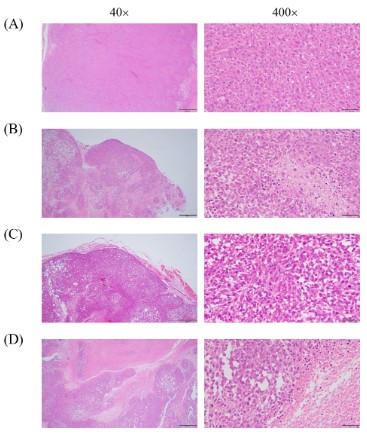
Pathomorphological changes in tumors
To further clarify the inhibitory effect of DHA on tumors, H&E
staining was used for the pathological analysis of tumor cells. H&E
staining results revealed that tumor cells in the model group grew
in like nests with low differentiation and a large number of nuclear divisions could be observed, as seen in Figure 2 (A). Tumor
cells in the cisplatin group grew in a nested pattern and necrosis
was visible in the tumor cell foci, which were smaller than those
in the model group, as shown in Figure 2 (B).
As shown in Figure 2 (C), the tumor cells in the low-dose DHA
group exhibited nest-like proliferation and nuclear division, as
well as little differentiation and necrosis. In the high-dose group,
the tumor cells developed in a nested pattern, and the cell foci
underwent a great deal of necrotic apoptosis, as seen in Figure
2 (D).
As evident from the results of H&E staining, indicating that DHA
impeded human CRC cancer cell growth, and induced apoptosis in
vivo. It provided a way for in vitro research on the mechanism.
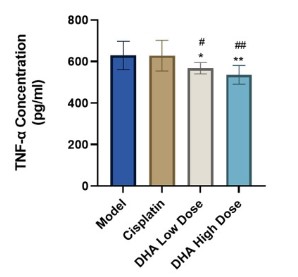
Effects of DHA on tumor TNF-α levels
Elisa was used to measure the amount of TNF-α in each group
of mice's peripheral blood. The results demonstrated that there
was no statistically significant difference between the cisplatin
group and the model group in serum TNF-α levels (P>0.05). The
content of TNF-α exhibited a statistically significant reduction in
both the high and low DHA dosage groups (P<0.05, P<0.01). Figure 3 demonstrated that when compared to the cisplatin group,
the inhibitory effect on TNF-α level was more substantial (P<0.05).
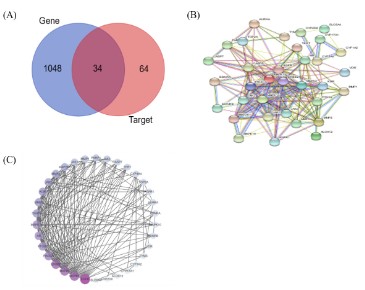
Targets identification and PPI network establishment
A total of 98 targets of DHA were obtained by SWISS and Sym-Map. GeneCards retrieval yielded a total of 21264 CRC-related
targets, and the top 1088 targets were chosen by the score in descending order. Drawing a Venn Diagram database allowed for the
matching analysis of the target of DHA and genes related to CRC,
and 34 overlap targets were chosen, as shown in Figure 4 (A), to
indicate the target of DHA for the treatment of CRC.
The 34 intersection targets of DHA-CRC were imported into the
String to obtain the protein interaction data (Figure 4 (B)), and
then imported into Cytoscape to optimize the construction and
analysis of the PPI network (Figure 4 (C)). The PPI network had an
average node degree of 10.82, 34 nodes (common target genes),
and 368 edges. And the targets of the DHA's anti-CRC effect were
represented by the nodes in the diagram. In Figure 4 (C), the correlation is better, the nodes are larger, the color is darker, the
degree is higher, and the overall score value is greater. Following
Cytoscape Network analysis, the degree value for each target was
determined, and the top 5 degrees were EGFR, MAPK1, MAPK3,
ERBB2, and PIK3CA.
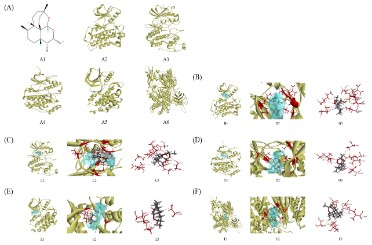
Molecular docking
For the purpose of molecular docking verification, DHA was
utilized as a ligand and the top 5 proteins in the PPI network-EGFR, MAPK1, MAPK3, ERBB2, and PIK3CA-were used as receptors. As shown in Figure 5, all 5 proteins can bind to small
molecules. The sequence of force strength was as follows:
MAPK3>MAPK1>ERBB2>EGFR >PIK3CA. According to the CDOC-KER INTERACTION ENERGY score, the molecule had a moderate
intensity interaction with the five proteins, ranging from 24 to 32.
TYR53, VAL56, ALA69, LYS71, LEU173, and CYS183 of the
MAPK3 protein all interacted with small molecules on an alkyl-alkyl basis. The small molecules could be effectively mixed in the
pocket and interacted with more amino acid residues despite the
weak contact force itself (Figure 5 (D)).
MAPK1's ILE31, VAL39, ALA52, LEU107, ASP111, LEU156, and CYS166 interacted with GLY32 via weakening hydrogen bonds and
alkyl-alkyl interactions with small molecules. Despite having larger contact forces, the MAPK1 protein's function was less powerful than the MAPK3 protein because of its greater distance (Figure
5 (C)).
The ERBB2 protein interacts with small molecules via four alkyl-alkyl interactions at LEU726, VAL734, and CYS806. It was evident that there weren't many forces acting, which made the binding weak (Figure 5 (E)).
The EGFR protein's VAL726, LSY745, and LEU844 interacted
with one another in a weak alkyl-alkyl-alkyl manner. It had weak
hydrogen bonding with ASP855 and strong hydrogen bonding
with LYS745, ARG841, ASN842, and THR854. The interaction was
weak despite the high force because the vast cavity of the protein
prevented small molecules of this shape from being bound to the
active site, despite the force being substantial (Figure 5 (B)).
The PIK3CA protein displayed mild alkyl interactions with
MET772, ILE848, and ILE932. Additionally, the PIK3CA protein's
SER774 and SER919 exhibited hydrogen bonds with small molecules, resulting in a weak overall interaction, which caused them
to be ranked last (Figure 5 (F)).
In conclusion, the top 5 primary targets that were strongly
related to DHA in the treatment of CRC were identified by Cytoscape's analysis of PPI Network data, and molecular docking was carried out to foretell the interaction between
receptor and ligand. These were the sequencing results:
MAPK3>MAPK1>ERBB2>EGFR>PIK3CA, indicating that DHA may
primarily regulate MAPK3, MAPK1, ERBB2, EGFR, and PIK3CA to
prevent CRC.
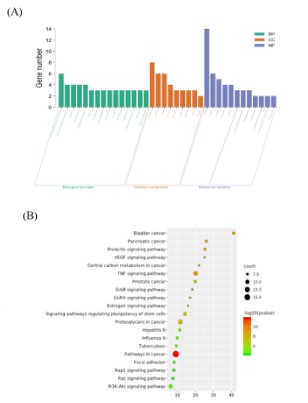
GO biological processes and KEGG enrichment pathway analysis
The GO function was applied to the target gene to clarify the
biological characteristics of the target. Figure 6 (A) displayed 15
BP enrichment analyses (including energy pathway and signal
transduction), while 9 CC and 12 MF enrichment analyses were
selected based on the count value. For specific items, see Table S1. The results of the GO biological process analysis suggested
that DHA might inhibit CRC by influencing a variety of biological
processes, including protein, protease, enzyme, heme, and iron
binding in the cell membrane, cytoplasm, mitochondria, receptor
complex, cytoskeleton, and extracellular matrix.
A KEGG pathway enrichment analysis was carried out to investigate the potential mechanism of action of DHA in the therapy of
CRC. The results revealed that 34 target genes were assigned to
86 KEGG pathways, and the top 20 pathways were then further
evaluated by the count value, as shown in Table S2. The analysis
results were then visualized by the micro message platform (Figure 6 (B)). The tumor signaling pathway had the highest number of target connections (count=17), and proteoglycan had 11
targets in cancer, followed by the PI3K/AKT signaling pathway
with 10 targets. PI3K/AKT involves multiple targets such as EGFR,
MAPK1, MAPK3, and PIK3CA, which were strongly linked to the
growth, differentiation, apoptosis, migration, and adhesion of tumor cells. Based on these results, it was theoretically conceivable
to treat CRC by utilizing DHA to prevent the PI3K/AKT signaling
pathway from becoming activated.
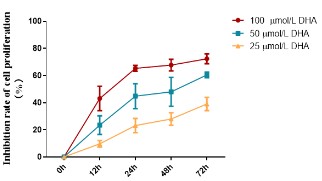
Effects of DHA on the proliferation of RKO cells
The equal concentration gradient established in the pre-experiment allowed us to determine the DHA IC50 value for RKO cells,
which was 94.14 μmol/L. Here, several concentrations (25, 50,
and 100 μmol/L) and periods (12, 24, 48, and 72 h) were set based
on the IC50 value to assess the influence of DHA on the survival of
RKO cells by MTT. This allowed us to analyze the anti-CRC activity
of DHA. Figure 7 illustrated how increasing the medication dosage
and duration of the intervention strengthened the inhibitory im-
pact of DHA on RKO cell growth.
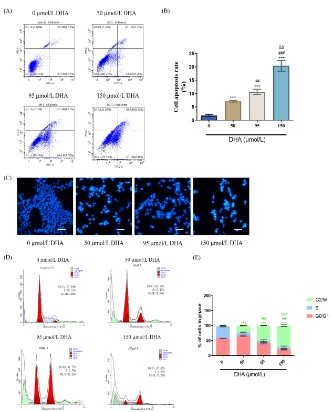
Effects of DHA on apoptosis and cycle of RKO cells
We investigated how DHA affected the apoptosis of RKO cells
using flow cytometry. The effects of different DHA concentrations
(50, 95, 150 μmol/L) on the apoptosis of RKO cells were depicted
in Figure 8 (A) and (B). The results indicated that, as compared to
the 0 μmol/L DHA group, DHA could considerably enhance apoptosis of RKO cells with an increased concentration within 48 h.
With different concentrations of DHA intervention, RKO cells were stained using Hoechst 33258 fluorescent dye. The apoptosis
of RKO cells induced by DHA intervention for 48 h was visible under a fluorescent microscope.
As seen in Figure 8(C), the fluorescence intensity gradually
deepened as the medication concentration rose, and the number
of cells declined. The nucleus ruptured and turned white at a high
concentration (150 μmol/L).
The cell cycle alterations in different dosages of DHA treated
with PI labeling were discovered using flow cytometry. Figures 8
(D) and (E) demonstrated how DHA damaged RKO cells’ DNA and
blocked the cell cycle in the G2/M phase, and higher DHA concentrations strengthened the degree of blockage. In Figure 8 (D), it
was also noted that the fraction of aggregation (green) and cell
debris (blue) increased as DHA concentration rose, indicating an
increase in the number of apoptotic cells. It proved that DHA prevented RKO cells from proliferation and inducing apoptosis.
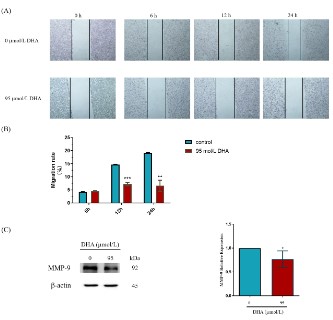
Effects of DHA on the migratory capacity of RKO cells
DHA was tested for its effects on the migration of RKO cells to
further confirm its anti-CRC activity. A wound healing experiment
was applied to examine the impact of 95 μmol/L DHA intervention at various times (6, 12, and 24 h) on the migratory capacity
of RKO cells, as shown in Figures 9 (A) and (B). When compared
to the control group (0 μmol/L DHA), DHA had no discernible impact on the RKO cells’ capacity for migration at 6 h. However, following 12 and 24 h of treatment, DHA was able to inhibit the sur-
vival of RKO cells and dramatically limit their migration (P<0.01).
Meanwhile, Figure 9 (C) showed that the addition of 95 μmol/L
DHA decreased the expression of MMP-9 in RKO cells (P<0.05).
These results implied that DHA suppressed RKO cell proliferation
and migration, which might have the potential application value
of the anti-CRC effect.
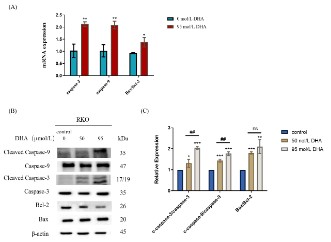
Effects of DHA on mRNA transcript levels of apoptosis-related
genes in RKO cells
We detected the activation of the caspase cascade by RT-qPCR
and western blot to better comprehend the impact of DHA on
RKO cell apoptosis and the molecular mechanism causing apoptosis. The major regulators for endogenous apoptotic pathways
are found in mitochondria, which also govern Cyt C release by
controlling the Bcl-2 protein family and the permeability of the
mitochondrial membrane. To create an "apoptotic body," released
Cyt C joins with Apaf-1 and pro-caspase-9. This "apoptotic body"
subsequently activates the executive factor caspase-3, resulting in
DNA damage and nuclear condensation, which leads to cell apoptosis [18,19]. In RKO cells, RT-qPCR was used to identify the mRNA
transcripts of pro-apoptotic factors including caspase-3, caspase-9, Bax, and anti-apoptotic factors like Bcl-2, shown in Figure
10 (A). After 12 h of 95 μmol/L DHA intervention, the expression
of caspase-3 and caspase-9 mRNA in RKO cells was considerably
higher than in the control group (0 μmol/L DHA), (P<0.01). The
Bax/Bcl-2 mRNA expression ratio was also increased (P<0.05).
Effects of DHA on the expression of apoptosis-related proteins in RKO cells
Figures 10 (B) and (C) indicated the changes in the expression
levels of apoptosis-related proteins in RKO cells after DHA intervention. The results demonstrated a considerable up-regulation
of cleaved-Caspase-3/Capase-3, cleaved-Caspase-9/Capase-9,
and Bax/Bcl-2 protein expression ratios in RKO cells following 48
h intervention with 50 and 95 μmol/L DHA. And compared to the
50 μmol/L DHA, 95 μmol/L DHA showed a more pronounced rise
in the expression of apoptosis-related proteins. These results suggested that DHA treatment of RKO cells could induce apoptosis
and prevent cell growth by activating the endogenous apoptotic
pathway.
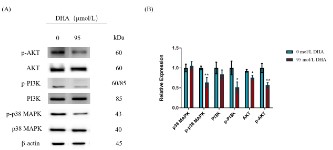
Effects of DHA on the phosphorylation of PI3K, AKT, and p38
MAPK in RKO cells
To support the underlying anti-CRC action of DHA that was discovered utilizing network pharmacology, the expression of proteins linked to the PI3K/AKT pathway was investigated. Western
blot was used to analyze the endogenous role of PI3K, AKT, and
p38 MAPK in the DHA intervention of RKO cells. As seen in Figure
11, 95 μmol/L DHA significantly reduced the protein expression of
AKT and prevented the phosphorylation of p38 MAPK, PI3K, and
AKT within 48 h when compared to the control group (0 μmol/L
DHA) (P<0.05, P<0.01). These results revealed a connection
between DHA's proapoptotic and proliferation-inhibiting effects
on RKO cells and the PI3K/AKT signal transduction pathway.
Discussion
The function and molecular mechanism of DHA in the treatment of CRC were investigated in this study using RKO cell lines,
and the suppressive impact of DHA on subcutaneous transplanted
tumors was evaluated using nude mice. The findings showed that
DHA could cause RKO cell apoptosis and decrease tumor growth
while not negatively affecting normal cells via blocking p38 MAPK
phosphorylation and modifying the PI3K/AKT signaling pathway
[20], indicating that DHA is a promising anticancer therapy.
To create the RKO CRC model so that researchers can examine
how different DHA concentrations affect CRC xenografts in nude
mice. The level of TNF-α expression in the serums of each group
after administration was found and examined. The weight of the
solid tumor was markedly smaller in the high-dose group than in
the model group and the low-dose group, suggesting that a tumor
may be suppressed by a high dose of 200 mg/kg DHA. The tumor
inhibition rate was 41.45%, whereas the corresponding percentage in the cisplatin group was 60.80%. Additionally, the body
weight and subcutaneous tumor volume changes in the mice before and after treatment revealed that while the cisplatin group
and the high-dose DHA group significantly inhibited the growth of
the transplanted tumor, the cisplatin group decreased the mice's
body weight and growth status.
On the other hand, inflammation and the immune system
considerably impact the occurrence and growth of tumors. In
distinct cellular microenvironments, TNF-α triggers a variety of
reactions, including the activation of apoptosis, necrosis, angiogenesis, cell migration, and differentiation. The NF-κB signaling
pathway is triggered when TNF-α interacts with the receptor
TNF-α, and NF-κB transcription factors cause the expression of apoptosis inhibitory factors, inhibiting the activation of caspases
and preventing tumor cell apoptosis, which promotes tumor
growth, invasion, and metastasis [21]. Clinical research findings
also revealed that blood samples from CRC patients had much
higher levels of TNF-α than samples from healthy individuals [22].
Elisa was used to measure the amount of TNF-α in each group of
mice's peripheral blood. Both the high- and low-dose DHA groups
dramatically decreased the amount of TNF-α, it was discovered.
In comparison to the cisplatin group, the inhibitory effect on the
level of TNF-α was more substantial (P<0.05).
The regulation of transcription, translation, proliferation,
growth, and survival, as well as other fundamental cell biological processes, is governed by the PI3K/AKT and MAPK signaling
pathways. These pathways have a key role in maintaining protein synthesis, fostering proliferation, and preventing apoptosis.
Consequently, inappropriate activation of this pathway can result
in the emergence and growth of malignancies [23,24]. PI3K is a
specific catalyst for phosphatidylinositol, primarily consisting of
its regulatory subunit P85 and its catalytic subunit P110. It can be
classified into subunits I, II, and III based on the catalytic subunit
P110 and its related substrates. According to studies, phosphatidylinositol PIP2 activates the plasma membrane's PI3K diphosphate function. This causes PIP2 to be transformed into PIP3,
which is subsequently transferred as a second messenger bound
to AKT [25].
The top 5 key targets that are strongly related to DHA in the
treatment of CRC were identified by the analysis of PPI Network
data by Cytoscape, and molecular docking was carried out to estimate the interaction between receptor and ligand. These findings
imply that DHA may inhibit the development of CRC via regulating
MAPK3, MAPK1, ERBB2, EGFR, and PIK3CA. PIK3CA participates in
the PI3K/AKT signaling pathway and encodes the p110α protein,
which is one of the PI3K enzyme's protein subunits. According to
studies, when PIK3CA co-mutates with other genes, the PI3K enzyme is continually activated, which results in the development
of tumors even though PIK3CA itself does not cause cancers [26].
The ERBB2 gene is also known as the HER2 gene. Numerous malignancies, including ovarian, bladder, and breast cancer, have
been linked to the overexpression of the ERBB2 gene, according
to studies [27-29]. ERBB2 can regulate the PI3K/AKT signaling pathway, which can impact the multiplication and invasion of tumor
cells. The PI3K/AKT signaling route predominantly boosts Ras proteins by dimerization, initiating the phosphorylation cascade. Like
ERBB2, EGFR is a key upstream of this signaling pathway [30].
Akt is a part of a signaling complex that also comprises p38
MAPK, MK2, and Hsp27. In human neutrophils, p38 MAPK-dependent MK2 activation functions as PDK2, which can control Akt
activity and is crucial for Akt phosphorylation [31]. The phosphorylation of PDK1, PDK2, and its associated gene sites leads to the
activation of AKT, which then controls cell division, death, and
proliferation. Through a series of anti-apoptotic effects, activated Akt can activate the phosphorylation of Bcl-2, caspase, and
FOXOs, ultimately boosting the survival of cancer cells [32,33].
The results showed that 95 μmol/L DHA completely reduced
the expression of the AKT protein in RKO cells and significantly
suppressed the phosphorylation of p38 MAPK, PI3K, and AKT,
while also increasing the levels of caspase-3, caspase-9, and Bax/Bcl-2 in RKO cells, thereby triggering the endogenous apoptosis
pathway. RKO cells' motility and viability may both be considerably hampered by DHA (P<0.01). Further evidence that DHA
therapy decreased the expression of MMP-9 protein in RKO cells
came from a western blot analysis (P<0.05). MMP-9 has been
shown to promote angiogenesis and the destruction of the ECM
and basement membrane in cancer cells [34]. Reducing MMP-9's activity in the PI3K/AKT pathway can boost the expression of
downstream apoptosis-related proteins because MMP-9 is a prominent molecule downstream of the process [35]. These findings
indicated that the link between DHA inhibition of RKO cell proliferation and migration as well as apoptosis induction depended on
the activities of p38 MAPK, PI3K, and AKT.
Funding: This work was supported by grants from the National Nature Science Foundation of China (No.82003641), the National Science and Technology Major Project (2019ZX09201004-002), and the Shanghai Education Commission budget project
(18LK019).
References
- Yang Q, Hou C, Huang D, Zhuang C, Jiang W, et al. miR-455-5p functions as a potential oncogene by targeting galectin-9 in colon cancer. Oncology letters. 2017; 13: 1958-1964.
- Li Y, Wu Y, Xia Q, Zhao Y, Zhao R, et al. Platycodon grandiflorus
enhances the effect of DDP against lung cancer by down regulating PI3K/Akt signaling pathway. Biomedicine & pharmacotherapy
= Biomedecine & pharmacotherapie. 2019; 120: 109496.
- Han JM, Song HY, Kim KI, Park WY, Park SH, et al. Polysaccharides
from Annona muricata leaves protect against cisplatin-induced
cytotoxicity in macrophages by alleviating mitochondrial dysfunction. Molecular medicine reports. 2023; 27.
- Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese journal of natural medicines. 2013; 11: 110-120.
- Hao da C, Xiao PG. Network pharmacology: a Rosetta Stone for
traditional Chinese medicine. Drug development research. 2014;
75: 299-312.
- Tong Y, Liu Y, Zheng H, Zheng L, Liu W, et al. Artemisinin and its
derivatives can significantly inhibit lung tumorigenesis and tumor
metastasis through Wnt/β-catenin signaling. Oncotarget. 2016; 7:
31413-31428
- Wang Z, Li M, Liu Y, Qiao Z, Bai T, et al. Dihydroartemisinin triggers
ferroptosis in primary liver cancer cells by promoting and unfolded
protein response-induced upregulation of CHAC1 expression. Oncology reports. 2021; 46
- Ma Y, Zhang P, Zhang Q, Wang X, Miao Q, et al. Erratum: Dihydroartemisinin suppresses proliferation, migration, the Wnt/β-catenin pathway and EMT via TNKS in gastric cancer. Oncology
letters. 2022; 23: 34.
- Cui W, Fang T, Duan Z, Xiang D, Wang Y, et al. Dihydroartemisinin
Sensitizes Esophageal Squamous Cell Carcinoma to Cisplatin by
Inhibiting Sonic Hedgehog Signaling. Frontiers in cell and developmental biology. 2020; 8: 596788.
- Yuan B, Liao F, Shi ZZ, Ren Y, Deng XL, et al. Dihydroartemisinin Inhibits the Proliferation, Colony Formation and Induces Ferroptosis of
Lung Cancer Cells by Inhibiting PRIM2/SLC7A11 Axis. OncoTargets
and therapy. 2020; 13: 10829-10840.
- Zhang T, Hu Y, Wang T, Cai P. Dihydroartemisinin inhibits the viability of cervical cancer cells by upregulating caveolin 1 and mitochondrial carrier homolog 2: Involvement of p53 activation and
NAD(P)H:quinone oxidoreductase 1 downregulation. International
journal of molecular medicine. 2017; 40: 21-30.
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, et al. PubChem Substance and Compound databases. Nucleic acids research. 2016; 44:
D1202-13.
- Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, et al.
SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic acids research. 2014; 42(Web Server
issue): W32-8.
- Wu Y, Zhang F, Yang K, Fang S, Bu D, et al. SymMap: an integrative
database of traditional Chinese medicine enhanced by symptom
mapping. Nucleic acids research. 2019; 47: D1110-d7.
- Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, et
al. Human Gene-Centric Databases at the Weizmann Institute of
Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic acids research. 2003; 31: 142-146.
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B.
STRING: a database of predicted functional associations between
proteins. Nucleic acids research. 2003; 31: 258-261.
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids
research. 2007; 35: W169-75.
- Yang Y, Yu Y, Wang J, Li Y, Li Y, et al. Silica nanoparticles induced
intrinsic apoptosis in neuroblastoma SH-SY5Y cells via CytC/Apaf-1
pathway. Environmental toxicology and pharmacology. 2017; 52:
161-169.
- Lu WN, Zheng FP, Lai DW, Li H. Xuezhikang () reduced renal cell
apoptosis in streptozocin-induced diabetic rats through regulation
of Bcl-2 family. Chinese journal of integrative medicine. 2016; 22:
611-618.
- Chen T, Li M, Zhang R, Wang H. Dihydroartemisinin induces apoptosis and sensitizes human ovarian cancer cells to carboplatin therapy. Journal of cellular and molecular medicine. 2009; 13: 1358-
1370.
- Baud V, Karin M. Is NF-kappaB a good target for cancer therapy?
Hopes and pitfalls. Nature reviews Drug discovery. 2009; 8: 33-40.
- Rasool M, Malik A, Waquar S, Ain QT, Rasool R, Asif M, et al. Assessment of clinical variables as predictive markers in the development and progression of colorectal cancer. Bioengineered. 2021;
12: 2288-2298.
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Montalto G,
et al. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/
PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget. 2012; 3: 954-987.
- Asati V, Mahapatra DK, Bharti SK. PI3K/Akt/mTOR and Ras/Raf/
MEK/ERK signaling pathways inhibitors as anticancer agents:
Structural and pharmacological perspectives. European journal of
medicinal chemistry. 2016; 109: 314-341.
- Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/
AKT pathway for cancer drug discovery. Nature reviews Drug discovery. 2005; 4: 988-1004.
- Xu JM, Wang Y, Wang YL, Wang Y, Liu T, et al. PIK3CA Mutations
Contribute to Acquired Cetuximab Resistance in Patients with
Metastatic Colorectal Cancer. Clinical cancer research : an official
journal of the American Association for Cancer Research. 2017; 23:
4602-4616.
- Ross JS, Gay LM, Wang K, Ali SM, Chumsri S, et al. Nonamplification
ERBB2 genomic alterations in 5605 cases of recurrent and metastatic breast cancer: An emerging opportunity for anti-HER2 targeted therapies. Cancer. 2016; 122: 2654-2662.
- Skrypek N, Vasseur R, Vincent A, Duchêne B, Van Seuningen I, et
al. The oncogenic receptor ErbB2 modulates gemcitabine and irinotecan/SN-38 chemoresistance of human pancreatic cancer cells
via hCNT1 transporter and multidrug-resistance associated protein MRP-2. Oncotarget. 2015; 6: 10853-10867.
- Yoshida K, Tsuda M, Matsumoto R, Semba S, Wang L, et al. Exosomes containing ErbB2/CRK induce vascular growth in premetastatic niches and promote metastasis of bladder cancer. Cancer
science. 2019; 110: 2119-2132.
- Kebenko M, Drenckhan A, Gros SJ, Jücker M, Grabinski N, et al.
ErbB2 signaling activates the Hedgehog pathway via PI3K-Akt in
human esophageal adenocarcinoma: identification of novel targets for concerted therapy concepts. Cellular signalling. 2015; 27:
373-381.
- Rane MJ, Coxon PY, Powell DW, Webster R, Klein JB, et al. p38 Kinase-dependent MAPKAPK-2 activation functions as 3-phosphoinositide-dependent kinase-2 for Akt in human neutrophils. The
Journal of biological chemistry. 2001; 276: 3517-3523.
- Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR Signaling in
Cancer. Frontiers in oncology. 2014; 4: 64.
- Huang WC, Hung MC. Induction of Akt activity by chemotherapy
confers acquired resistance. Journal of the Formosan Medical Association = Taiwan yi zhi. 2009; 108: 180-194.
- Lin W, Xie J, Xu N, Huang L, Xu A, et al. Glaucocalyxin A induces
G2/M cell cycle arrest and apoptosis through the PI3K/Akt pathway in human bladder cancer cells. International journal of biological sciences. 2018; 14: 418-426
- Li C, Qin Y, Zhong Y, Qin Y, Wei Y, et al. Fentanyl inhibits the progression of gastric cancer through the suppression of MMP-9 via
the PI3K/Akt signaling pathway. Annals of translational medicine.
2020; 8: 118.