Introduction
Lung cancer is the most common cancer in the world, and lung
squamous cell carcinoma (LUSC) is a major subtype of non-small
cell lung cancer with extremely poor prognosis [1,2]. Although the
treatment of lung cancer has made rapid progress, the treatment
of lung squamous cell carcinoma is limited, and the drive gene
mutation rate of lung squamous cell carcinoma is low [3]. In addition to 61 increasing the burden of patients, gene testing for
patients with squamous cell carcinoma has no prominent significance in guiding clinical treatment. Therefore, the guidelines do
not recommend gene testing as a routine examination for patients
with squamous cell carcinoma. In addition to routine surgery and
chemotherapy, immune checkpoint inhibitors have become an
available treatment for advanced squamous cell carcinoma of the
lung [4]. Therefore, there is an urgent need to find possible target
genes and efficacy evaluation indicators of immunotherapy.
The ubiquitin E3 ligase ITCH, also named atrophin-1 interacting
protein 4 (AIP4), is a member of the NEDD4 HECT-type family E3
ligases [5], which is an important member of the classic E3 ubiquitination process. In past studies, the role of ITCH in cancer has
been gradually revealed.
Depending on the difference of its ubiquitination substrate, it
may play a different role in cancer.
For example, ITCH overexpression has been observed in some
human cancers, including anaplastic thyroid carcinoma [6], breast
cancer and ovarian cancer [7]. However, in other studies, ITCH
plays an anti-tumor role, for example, its ubiquitin PD-L1 promotes its interpretation in melanoma [8], which may be due to
the wide substrate differences of ITCH. In addition to its effect
on tumor cells, ITCH also plays a regulatory role on immune cells
and tumor-related immune microenvironment. For example, ITCH
has played an active role in promoting the differentiation of Treg
[9,10]. However, clinical studies have shown that tumor infiltrating Tregs are usually associated with poor prognosis of cancer,
including lung cancer11. In addition to its effect on Treg cells, ITCH
can also inhibit the activation of T cell antigen receptor (TCR),
which is an important way for T cells to exert anti-tumor effect
[12]. In lung cancer, previous studies have shown that in lung adenocarcinoma, ITCH promotes the metastasis of lung cancer cells
under hypoxia [13], but 82 there is no report of ITCH function in
lung squamous cell carcinoma.
In this study, we first found that the expression of ITCH in lung
squamous cell carcinoma was significantly higher than that in
paracancerous tissue, and the number of ITCH copies in cancer
tissue was significantly increased, and its expression gradually
increased with the increase of stage, and the prognosis of lung
squamous cell carcinoma patients with high expression of ITCH
was poor. Based on these results, we further analyzed the correlation between ITCH and the clinicopathological characteristics
of lung squamous cell carcinoma, and found that it was significantly related to "aneuploid" and "Hyposia", which may be the
reason why ITCH promoted the progression of lung squamous
cell carcinoma. In addition, we found that the methylation of 91
ITCH promoter increased significantly, which may be an important
reason for the high expression of ITCH. Through pathway enrichment analysis, we explored the reason why ITCH promoted the
progression of lung squamous cell carcinoma. The results showed that ITCH was closely related to the metastasis process, and the
results of in vitro experiments also confirmed that ITCH could promote the metastasis of lung squamous cell carcinoma cell lines.
In addition to its effect on lung cancer cells, ITCH can reshape the
tumor microenvironment, induce the infiltration of M2 macrophages and neutrophils, and has a high immune score in patients
with low expression of ITCH lung squamous cell carcinoma, and is
sensitive to the effect of immune checkpoint 99 inhibitors. These
results preliminarily reveal the role of ITCH in lung squamous cell
carcinoma, and provide a basis for clinical prognosis and drug use.
Materials and methods
Data download and processing
TCGA database is an extremely important cancer genomics
project. We download sequencing data and clinical data of lung
squamous cell carcinoma through TCGA database (www.tcga-data. nci.nih. gov/tcga) [14]. The selected samples include ITCH gene
expression data and relevant clinical data, including age, sex, T, N,
M stages, pathological stages, survival status, etc. Since all data
are downloaded from the public database, the study does not require the approval of the Ethics Committee.
Gene expression analysis of ITCH
ITCH mRNA expression level in different cancer types were obtained from TIMER (http://timer.comp-genomics.org/) database
[15]. ITCH protein expression in different patient was also obtained from HPA database (https://www.proteinatlas.org/) [16]. In
order to detect the difference in the expression of ITCH in different Stages, N stages, and whether smokers or not, the TCGA-LUSC
data set was selected for detection using the UALCAN online data
set (https://ualcan.path.uab.edu/) [17]. Tnmplot online data set
(https://www.tnmplot.com/) [18] was used to detect the expression level of ITCH in tumor, adjacent tissues and metastatic lesions.
Prognostic analysis
First of all, R package was used to carry out univariate analysis
and umultivariate analysis on ITCH and pathological parameters,
and the analysis results are shown in heat map and forest map.
Kaplan-Meier curve was further used to analyze the difference in
survival rate between high and low expression of ITCH.
The nomogram was established by R “rms” and “survival” packages. Calibration curves were further used to assess the accuracy
of nomograms in differentiating patient groups.
Tumor Mutational Burden (TMB)
Download the RNA sequencing data, gene mutation data, and
clinical data of LUSC from the TCGA database (https://gdc-portal.
nci.nih.gov/). Data is stored in mutation annotation format and
processed with VarScan software. Use the "maftools" package to
analyze and summarize these mutation data [19]. After obtaining
the TMB value, we further analyzed the correlation between different ITCH expressions and the TMB value of each sample.
Correlation analysis of tumor immune infiltrating cells
CIBERSORT algorithm is used to quantify the proportion of immune cells in the mixed cell population [20,21]. RNA-Seq (FPKM
format) sample analysis can obtain each example of 22 immune
cell abundance matrix, including macrophages (macrophages M1, M2 macrophages and M0 macrophages), T cells (T cell helper (Tfh)
cells, memory CD4+T cells, and activated memory CD4+T cells, γδ
T cells, CD8+T cell subsets, and CD4+T cells), resting natural killer
(NK) cells, activated NK cells, mast cells, activated memory B
cells, resting dendritic cells, activated dendritic cells, naïve B cells,
monocytes, plasma cells, neutrophils, and eosinophils [21]. The
CIBERSORT result of the sample (p<0.05) shows that the immune
cell group fraction generated by CIBERSORT is accurate and can
be used for further analysis. Normalize the CIBERSORT output estimation and sum the immune cell type score to 1. Using the normalized gene expression data of TCGA-LUSC data set, the relative
proportion of 22 immune cell subtypes was determined using the
R "CIBERSORT" package to evaluate the correlation between the
expression of ITCH in TCGA-LUSC samples and the immune infiltrating cells.
Immunotherapy response prediction
Immune cell abundance identifier (ImmuCellAI) is a calculation
method, which was released in 2020 and is used to predict the
response to immune checkpoint blockade based on the abundance of immune cells (especially different T cell subsets) [22].
The abundance of infiltrating immune cells was calculated by ImmuCellAI and used to establish a reaction prediction model. The
radial basis function kernel support vector machine (RBF kernel
support vector machine) was used to establish the immune treatment response prediction model. The Cancer Immunome Atlas
(TCIA) provides the comprehensive immune genome analysis results of the next generation sequencing data (NGS) of 20 solid cancers from the cancer genome atlas (TCGA) and other data sources.
The cancer immunity atlas (TCIA) online tool is used to predict the
response of immunochemotherapy [23]. The quantitative score of
tumor immunogenicity ranges from 0 to 10, which is called immunophenotypic score (IPS). IPS can be used to predict the reaction
of immunosuppressive agents at immune checkpoints [23].
CNV and promoter methylation prediction
GSCA online portal (http://bioinfo.life.hust.edu.cn/GSCA/#/)
[24] was used to detect the change of ITCH gene CNV and the difference of promoter methylation sites, and detect the correlation
with the expression of ITCH mRNA, selecting TCGA-LUSC data set.
ITCH co-expression networks and pathway enrichment
The LinkedOmics database (http://www.linkedomics.org/login.php) [25] was used to determine the ITCH co-expression genes
by using Pearson’s correlation coefficient, filtering by TCGA-LUSC
dataset, the results were showed via volcanic map and heat maps.
Then, the Gene Ontology Biological Process (GO_BP), and KEGG
pathways of ITCH and its co-expression genes by using gene set
enrichment analysis (GSEA).
Cell lines and culture condition
The human lung cancer cell line H1563 was obtained from
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). H1563 was maintained in RPMI 1640 medium
containing 10% heat-inactivated foetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 mg/ml) under a mixture
of 95% air and 5% CO2. According to the cell 179 doubling time,
the average passage is 3-4 days.
SiRNA transfection
SiRNAs and negative control (NC) were purchased from View
Solid Biotech (Beijing, China). All siRNAs were transfected into
cells using jet PRIME reagent (Polyplus) according to the manufacturer’s protocol.
RNA extraction and quantitative reverse transcription realtime polymerase chain reaction (qRT-PCR)
Total RNA extracted as described above [26]. Primer sequences
for ITCH: Forward 189 (5ʹ-GACCGGCTGCCATCTTAGTC-3ʹ), Reverse
(5ʹ-GGGTTAAGGCGTTGTCTCCA-3ʹ).
Western blotting analysis
Western blotting was performed as our previously described [26]. The primary antibodies: Rabbit monoclonalanti-ITCH
(ab108515,1:500) was purchased from Abcam (USA). Rabbit monoclonal anti-GAPDH (5174S,1:2000) was purchased from Cell Signaling Technology (USA).
Migration and invasion assay
Transwell assays were performed to assess cell migration and
invasion ability as described [26]. In general, we knock down ITCH
in lung cancer cells, and the 24-wellchamber and polycarbonate
inserts with 8-μmpore size membranes (Corning, USA) was used
to carry outmigration assays.
The cells (7 × 104 cells/well) transfed with siRNA were loaded
into the upper chamber with 200 μl serum-free RPMI 1640 medium. The lower chambers contained 500 μl of RPMI 1640 with
10% FBS. After 48 hours, the migrated cells on to the outer side
of the membrane were stained with 0.1% Giemsa solution for 1h.
Five different fields were captured and counted at ×20 magnificationper well.
Wound healing assays
The treated cells are inoculated into a six hole plate. When the
cell confluence reaches about 90%, 200 μl pipette tip was used to
scratch a straight line. After that, the supernatant was replaced
with 209 fresh medium without FBS twice. The migration images
were taken 0 and 24 hours after the 210 scratch. All experiments
were repeated three times. Image J software was used to analyze
and quantify blank areas.
Statistical analysis
R software (https://r-forge.r-project.org/3.6.3) was used for
statistical analysis. The survival curve was compared by Kaplan-Meier method, and the log-rank test was used to evaluate the
statistical significance of the survival rate among the groups. For
unmatched samples, the comparison between the two groups
was analyzed by Mann-Whitney test or Student's t test. For paired samples, Wilcoxon rank sum test was used. Kruskal Wallis rank
sum test was used for multiple comparisons to compare the differences between groups. Pearson correlation analysis is used to
measure the degree of correlation between some variables, and
the correlation strength is determined by the value of correlation
coefficient r. In order to estimate the influence of single continuous gene expression variable and independent prognostic factors on prognosis, single factor and multiple factor Cox regression
analysis were performed respectively. P value <0.05 is considered statistically significant.
Results
ITCH is highly expressed in lung squamous cell carcinoma
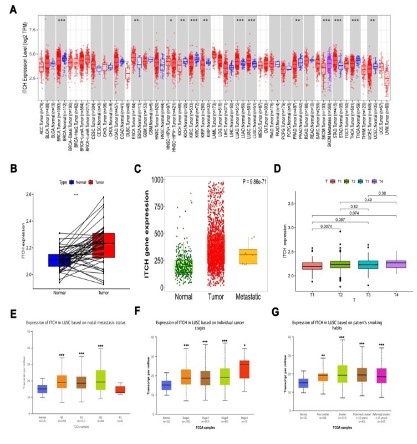
First, the TIMER online data set was used to detect the expression level of ITCH gene in various tumors and paracancerous tissues. The results showed that the expression level of ITCH gene
in lung cancer tissue was significantly higher than that in paracancerous tissues. Both lung adenocarcinoma and lung squamous
cell carcinoma had statistical significance, but the expression difference in lung squamous cell carcinoma was greater (Figure 1A).
We further downloaded TCGA-LUSC to further verify the difference between its expression in cancer and paracancerous tissues.
The results were consistent with our expectations. The expression
of ITCH in lung squamous cell carcinoma was significantly higher
than that in paracancerous tissues (Figure 1B), and the expression was higher in metastatic lesions (Figure 1C). Further analysis
of the correlation between its expression and clinicopathological
parameters showed that the expression of ITCH was statistically
different only from T1 to T2 in T stage. Although the expression of
ITCH was 239 higher in T4, there was no statistical difference compared with T1, which suggested that ITCH might play an important
role in the formation of early local lesions (Figure 1D).
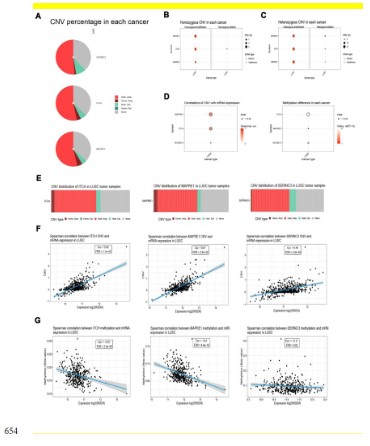
Increased copy number and decreased promoter methylation
of ITCH in lung squamous cell carcinoma
We further explored the factors that increase the expression of
ITCH in lung squamous cell carcinoma. At the same time, we analyzed the two genes that have the highest correlation with ITCH,
SERNIC3 and MAPRE1. The results showed that the amplification
of these three genes is the main change in lung squamous cell carcinoma. Among them, hete.amp is the important way, homo.amp
accounts for a small percentage, and the deletion part accounts
for a small part (Figure 3A). Figures 3B and 3C show the details
of different amplification methods. Figure 3E shows the specific
changes of copy types of these three genes.Further, we evaluated whether this copy number amplification led to the change of
mRNA expression. The results showed that the change of copy
number of ITCH and MAPRE1 was significantly positively correlated with mRNA 269 expression, with a correlation coefficient
greater than 0.5, but this correlation was less obvious on the SER-NIC3 gene, with a correlation coefficient of 0.48 (Figures 3D,F).
Figure 3F shows the specific correlation coefficients and statistical
differences. In addition, we detected the promoter methylation
status and mRNA expression of these three genes, and found that the methylation level of ITCH and MAPRE1 promoters was
significantly negatively correlated with their mRNA expression.
Figure 3G shows the specific correlation coefficient and statistical
difference. These results led us to speculate that the high expression of ITCH may be due to the abnormal amplification of its copy
number and the reduction of methylation in its promoter.
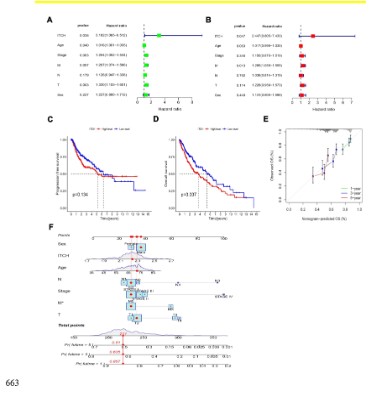
ITCH is a prognostic indicator of lung squamous cell carcinoma
Next, we focus on evaluating the prognostic predictive role and
applicability of ITCH in lung 280 squamous cell carcinoma. Based
on univariate analysis (Figure 4A) of prognostic factors and umultivariate analysis (Figure 4B), we found that ITCH is a stable adverse
prognostic factor for lung squamous cell carcinoma. In univariate
analysis, age/stage/T/M stage and the expression of ITCH were
both adverse prognostic factors of lung squamous cell carcinoma,
and both were statistically significant. However, in multivariate
analysis, only M stage and the expression of ITCH were statistically significant, indicating that ITCH was a more stableindependent prognostic indicator of lung squamous cell carcinoma than
clinicopathological parameters. Finally, we examined the role of
ITCH in the prognosis of lung squamous cell carcinoma, and found
that patients with high expression of ITCH had poor overall prognosis (Figure 4D). But PFS has no statistical significance (Figure
4C). Based on this result, we built a diagnostic nomogram predicting the 1 -, 3 -, and 5-year OS (Figure 4F). Draw calibration curve
to prove the prediction effectiveness of OS in the constructed nomograph (Figure 4E). These results suggest that ITCH is a relatively stable independent prognostic factor for lung squamous cell
carcinoma and can help predict survival time.
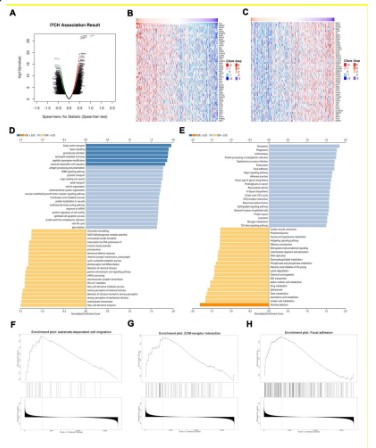
ITCH participates in the metastasis of lung squamous cell carcinoma
In order to understand the biological function of ITCH in lung
squamous cell carcinoma, we explored the co-expression pattern
of ITCH in TCGA-LUSC by applying the LinkFinder module in LinkedOmics online website. First of all, ITCH-related genes in lung
squamous cell carcinoma were detected and expressed by volcanic map (Figure 5A). Figures 5B and 5C show the heat map of
the first 50 genes with positive and negative correlation with ITCH
respectively. In addition, based on these differentially expressed
genes, the GO and KEGG analysis was carried out to evaluate the
possible way of ITCH to play its biological function. The results
showed that the GO term annotation (Figure 5D) showed that the
co-expressed genes of ITCH were mainly involved in "substrate-dependent cell migration" (Figure 5F). KEGG pathway analysis
(Figures 5E) shows that ITCH may participate in the "ECM-receptor interaction" (Figure 5G) and "Focal adhesion" (Figure 5H) pathway. These results showed that ITCH expression network had a
great influence on cell adhesion and metastasis of lung squamous
cell carcinoma. These results suggest that ITCH may participate in
the process of cell metastasis of lung squamous cell carcinoma,
thus promoting the progression of lung squamous cell carcinoma.
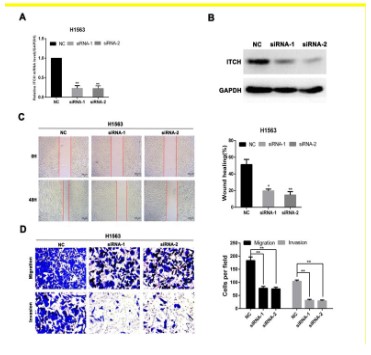
Therefore, we will test the metastasis effect of ITCH on lung squamous cell carcinoma cells in vitro. Figures 6A and 6B show the
changes of mRNA and protein levels of ITCH knockdown in lung
cancer cells. The ability of cell migration and invasion was significantly down-regulated after the knockout of ITCH (Figure 6C),
and the wound healing experiment also showed that the ability of
cell migration was also down-regulated after the knockout of ITCH
(Figure 6D). This is consistent with our previous online analysis
results, indicating that ITCH is involved in the metastasis process
of lung squamous cell carcinoma, thus promoting the progression
of lung squamous cell carcinoma and affecting its prognosis.
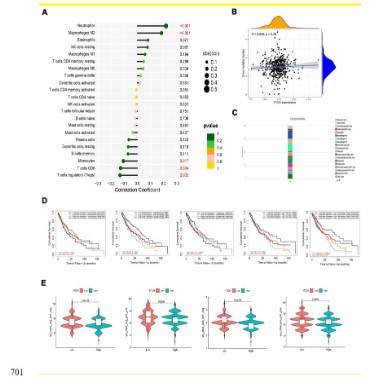
The correlation between ITCH and immune cell infiltration
and the efficacy of guiding immune checkpoints
We further tested the role of ITCH in the infiltration of immune
cells. The results showed that the high expression of ITCH was
in direct proportion to the infiltration of M2 macrophages and
neutrophils and had statistical significance, while it was in inverse
proportion to CD8+T cells, Tregs cells and mono cells and had
statistical significance (Figure 7A). However, the tumor mutation
load (TMB) related to the efficacy index of the current immune
checkpoint inhibitor has no obvious correlation (Figure 7B). The
single-cell data further refined the cell composition of the sample,
and found that the largest component of cells is macrophages, of
which M2 and M0 type account for the majority (Figure 7C). In addition, we analyzed the prognosis of these immune cell types with
strong correlation, and found that the difference in infiltration of
other immune cell subtypes, except neutrophils, would cause
the difference in prognosis, of which the more obvious impact is
macrophages, indicating that ITCH may further promote the progression of lung squamous cell carcinoma through the infiltration
and polarization of macrophages cells (Figure 7D). Based on the
correlation between ITCH and immune cells, we further tested the differences of IPS scores based on PD1 and CTLA4 expression
in different expression of ITCH samples. The results showed that
compared with the low expression group, IPS scores in the high
expression of ITCH group were lower than those in the low expression group, which indicated that the efficacy of using immune
checkpoint inhibitors in the high expression of ITCH lung squamous cell carcinoma might be average (Figure 7E), which provided
basic guidance for clinical guidance of the application of immune
checkpoint inhibitors.
Discussion
Most of the functions of ITCH depend on ITCH-deficient mice,
which play an important role in the regulation of immune system
as ubiquitin ligase. Lack of ITCH in mice has been proved to cause
serious autoimmune disease [27]. In human body, ITCH can also
cause immune deficiency syndrome [28]. This may be due to the
fact that ITCH down-regulates the inflammatory signal pathway
through ubiquitin, especially the T cell-mediated immune response [29,30]. However, in tumors, ITCH inhibits the activation
of T cell antigen receptor (TCR), thus inhibiting T cell activity
[12,31]. In our study, we also reached a similar conclusion that
high expression of ITCH is negatively correlated with CD8+T cell
infiltration, which may a mechanism of ITCH promoting tumor
progression. However, ITCH is inversely correlated with M2 type
macrophages and Treg cells. There is a significant positive correlation with M2 type macrophages that promote cancer, but there is
a negative correlation with Treg cells that promote cancer [32,33].
This may be related to different inflammation-related ubiquitination substrates of ITCH. For example, recent studies have found
that ITCH binds, ubiquitinates and downregulates PD-L1/L2 on
the tumor surface in human melanoma cells treated with MAPKi,
So as to promote T cell activation [8]. This may be caused by the
difference of expression position of ITCH in cells, but the specific
mechanism needs further exploration. In addition, the prognosis
analysis based on different immune cell infiltration showed that
macrophages, CD8+T and Treg cells had a significant impact on
the prognosis, indicating that ITCH might promote tumor progression by reshaping the immune microenvironment and readjusting
the proportion of immune-related cell infiltration. Based on this,
we further analyzed the efficacy of immune checkpoint inhibitors, and found that the immune score in the low expression ITCH
group was higher, and it might be easier to benefit from immune
checkpoint inhibitors.
For the evaluation of efficacy sensitivity of immune checkpoints, we also evaluated the relationship between the expression of ITCH and aneuploid score. Aneuploid is an imbalance in
the number of chromosomes or chromosome arms, which is an
almost universal feature of human cancer. Some studies have revealed that the negative effect of tumor aneuploidy on anti-tumor
immunity may be through the mechanism of immune evasion,
including down-regulation of PD-L1 expression and inhibition of
CD8+T cell reaction in tumor [34,35]. In addition, previous work
has determined that the increase of aneuploidy in tumor is a marker of low overall survival rate, and suggested that aneuploidy be
used as a biomarker of clinical results [36]. Recently, it has also
been found that aneuploidy is a powerful predictor of survival in
patients with non-small cell lung cancer receiving immunotherapy
[37], which is also consistent with our results. In lung squamous
cell carcinoma with high expression of ITCH, the aneuploid score
is higher, which indicates a poor prognosis. Research shows that
patients with high aneuploid scores may be more likely to benefit
from immune checkpoint inhibitors [38,39]. Combined with our
analysis results, that is, patients with lung squamous cell carcinoma with high expression of ITCH are likely to benefit from immune
checkpoint inhibitors, but this may contradict the efficacy indicators of immune checkpoint inhibitors obtained from the expression of PD-1 and CTLA4. The other approved indicator that can
be used to guide the application of immunosuppressive agents
at immune detection points is TMB [40]. Our research shows that
there is a weak positive correlation between the expression of
TMB and the expression of ITCH. Previous literature also shows
that the correlation between TMB and aneuploid score is very
small [39]. Therefore, this may also explain why the expression
of ITCH is significantly correlated with aneuploid score, but the
correlation with TMB is not very obvious, which also shows that
the prognostic significance of TMB in guiding immune checkpoint
therapy in lung squamous cell carcinoma is very limited. As for
the contradictory view that the immune efficacy score shows that
patients with low expression of ITCH lung squamous cell carcinoma may benefit from immunotherapy while the aneuploid score
shows that patients with high expression of ITCH lung squamous
cell carcinoma may benefit from immunotherapy, we prefer the
prediction result of aneuploid score, Because more and more
studies have shown that ITCH directly regulates the expression of
PD-L1 in tumor cells [8], it may cause a deviation in the score. In
addition to its role in immunity, the role of ITCH in tumor cells has
been gradually revealed.
For example, in breast cancer, ITCH can negatively regulate tumor suppressor LATS1 through ubiquitination, thereby increasing
YAP activity, and activating Hippo kinase pathway, leading to increased cell proliferation, decreased apoptosis, and promoting
tumor cell EMT [41]. In triple negative breast cancer (TNBC), the
nuclear localization of ITCH makes TNBC cells have DDR inhibition
to counteract replication stress and increase the survival rate and
growth potential of cancer cells [42]. In lung adenocarcinoma,
ITCH promotes the metastasis of lung cancer cells under hypoxia
[13]. In this study, we focused on the treatment resistance subgroup of lung squamous cell carcinoma. The results showed that
ITCH was highly expressed in lung squamous cell carcinoma, and
the prognosis of patients with high expression of ITCH was poor.
In terms of mechanism, our results were consistent with previous
research results. ITCH could promote the metastasis of lung squamous cell carcinoma cells. In the analysis of pathway enrichment,
we also found that ITCH could regulate the Hippo pathway in lung
squamous cell carcinoma, This is consistent with previous research results [43,44]. However, whether it can promote cancer
through the same pathway 407 in lung squamous cell carcinoma
has not been verified, which is also our next experimental plan.
In view of the important role of ITCH in lung squamous cell carcinoma, we also explored the possible reasons for the imbalance
of ITCH expression in lung squamous cell carcinoma. The 410 analysis results showed that the mRNA expression level of ITCH was
significantly correlated with the copy number (CNV) and methylation level. The increase of the amplified copy number led to the
increase of ITCH expression, but methylation often inhibited the
expression of target genes [45].
This trend also existed in this study, The expression of ITCH is
significantly related to the level of methylation, which makes us
speculate whether the decreased methylation of the promoter
site of ITCH leads to a significant increase in the expression of
ITCH, which needs further validation in vitro and in vivo
Conclusion
Based on these results, we conclude that the expression of
ITCH is increased in lung squamous cell carcinoma, and promotes
the metastasis of lung squamous cell carcinoma, leading to poor
prognosis. At the same time, the highly expressed ITCH regulates
the redistribution of tumor immune cells by reshaping the tumor
microenvironment, and promotes the immune escape of tumor
cells. In addition, its high expression may be due to the methylation imbalance of its promoter site. More importantly, ITCH
can be used as an important indicator of the benefits of clinical
application of immune checkpoints, providing a basis for clinical
guidance of drug use.
Declarations
Conflict of interest: The authors have declared that no competing interest exists.
Data availability statement: The data that support the findings
of this study are available from the First author, upon reasonable
request.
Funding statement: There is no funding statement in this study.
Conflict of interest disclosure: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict
of interest.
Author contributions: G-LB and G-JF conceived and designed
the study. Online data download and analysis by G-JF. G-LB did the
experiments and analyzed results, and written the manuscript.
G-LB and G-JF conducted experimental guidance. G-JF revised
the manuscript.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for
36 Cancers in 185 Countries. CA: a cancer journal for clinicians.
2021; 71: 209-249.
- Pan Y, Han H, Labbe KE, Zhang H, Wong KK. Recent advances in pre-clinical models for lung 447 squamous cell carcinoma. Oncogene.
2021; 40: 2817-2829.
- Kim Y, Hammerman PS, Kim J, et al. Integrative and comparative
genomic analysis of lung squamous cell carcinomas in East Asian
patients. Journal of clinical oncology : official journal 450 of the
American Society of Clinical Oncology. 2014; 32: 121-128.
- Paik PK, Pillai RN, Lathan CS, Velasco SA, Papadimitrakopoulou V.
New Treatment Options in Advanced Squamous Cell Lung Cancer.
American Society of Clinical Oncology educational book. American Society of Clinical Oncology. Annual Meeting. 2019; 39: e198-e206.
- Rotin D, Kumar S. Physiological functions of the HECT family of
ubiquitin ligases. Nature reviews. Molecular cell biology. 2009; 10:
398-409.
- Ishihara T, Tsuda H, Hotta A, et al. ITCH is a putative target for a
novel 20q11.22 amplification detected in anaplastic thyroid carcinoma cells by array-based comparative genomic hybridization.
Cancer science. 2008; 99: 1940-1949.
- Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo
pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer research. 2011; 71: 2010-2020.
- Yang Z, Wang Y, Liu S, et al. Enhancing PD-L1 Degradation by ITCH
during MAPK Inhibitor 462 Therapy Suppresses Acquired Resistance. Cancer discovery. 2022; 12: 1942-1959.
- Venuprasad K, Huang H, Harada Y, et al. The E3 ubiquitin ligase
Itch regulates expression of transcription factor Foxp3 and airway
inflammation by enhancing the function of transcription factor
TIEG1. Nature immunology. 2008; 9: 245-253.
- Peng DJ, Zeng M, Muromoto R, et al. Noncanonical K27-linked polyubiquitination of TIEG1 regulates Foxp3 expression and tumor
growth. Journal of immunology (Baltimore, Md.: 468 1950). 2011;
186: 5638-5647.
- Petersen RP, Campa MJ, Sperlazza J, et al. Tumor infiltrating Foxp3+
regulatory T-cells are associated with recurrence in pathologic
stage I NSCLC patients. Cancer. 2006; 107: 2866-2872.
- Huang H, Jeon MS, Liao L, et al. K33-linked polyubiquitination of T
cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010; 33: 60-70.
- Sun Q, Wang BB, Wei W, et al. ITCH facilitates proteasomal degradation of TXNIP in hypoxia- induced lung cancer cells. Thoracic
cancer. 2022; 13: 2235-2247.
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An 477 immeasurable source of knowledge. Contemporary oncology (Poznan, Poland). 2015; 19: A68-77.
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer research.
2017; 77: e108-e110.
- Navani S. Manual evaluation of tissue microarrays in a high-throughput research project: The 482 contribution of Indian surgical pathology to the Human Protein Atlas (HPA) project. Proteomics. 2016; 16: 1266-1270.
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN:
A Portal for Facilitating 485 Tumor Subgroup Gene Expression and
Survival Analyses. Neoplasia (New York, N.Y.). 2017; 19: 649-658.
- Bartha Á, Győrffy B. TNMplot.com: A Web Tool for the Comparison
of Gene Expression in 488 Normal, Tumor and Metastatic Tissues.
International journal of molecular sciences. 2021; 22.
- Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools:
efficient and comprehensive analysis of somatic variants in cancer.
Genome research. 2018; 28: 1747-1756.
- Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods in
molecular biology (Clifton, N.J.). 2018; 1711: 243-259.
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell
subsets from tissue expression profiles. Nature methods. 2015;
12: 453-457.
- Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: A Unique Method for
Comprehensive T-Cell 498 Subsets Abundance Prediction and its
Application in Cancer Immunotherapy. Advanced science (Wein-heim, Baden-Wurttemberg, Germany). 2020; 7: 1902880.
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell
reports. 2017; 18: 248-262.
- Liu CJ, Hu FF, Xie GY, et al. GSCA: An integrated platform for gene
set cancer analysis at 504 genomic, pharmacogenomic and immunogenomic levels. Briefings in bioinformatics. 2023; 24.
- Vasaikar SV, Straub P, Wang J, Zhang B. Linked Omics: Analyzing
multi-omics data within and 507 across 32 cancer types. Nucleic
acids research. 2018; 46: D956-d963.
- Gong LB, Wen T, Li Z, et al. DYNC1I1 Promotes the Proliferation and
Migration of Gastric 509 Cancer by Up-Regulating IL-6 Expression.
Frontiers in oncology. 2019; 9: 491.
- Lohr NJ, Molleston JP, Strauss KA, et al. Human ITCH E3 ubiquitin
ligase deficiency causes syndromic multisystem autoimmune disease. American journal of human genetics. 2010; 86: 447-453.
- Patel T, Henrickson SE, Moser EK, et al. Immune Dysregulation in
Human ITCH Deficiency Successfully Treated with Hematopoietic
Cell Transplantation. The journal of allergy and clinical immunology. In practice. 2021; 9: 2885-2893.e2883.
- Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nature
immunology. 2004; 5: 883-890.
- Matesic LE, Copeland NG, Jenkins NA. Itchy mice: the identification
of a new pathway for the development of autoimmunity. Current
topics in microbiology and immunology. 2008; 321: 185-200.
- Courtney AH, Lo WL, Weiss A. TCR Signaling: Mechanisms of Initiation and Propagation. Trends in biochemical sciences. 2018; 43:
108-123.
- Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers.
2014; 6: 1670-1690.
- Kryczek I, Wei S, Zou L, et al. Cutting edge: Th17 and regulatory T
cell dynamics and the regulation by IL-2 in the tumor microenvironment. Journal of immunology (Baltimore, Md. : 1950). 2007;
178: 6730-6733.
- Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches
to Understanding Cancer Aneuploidy. Cancer cell. 2018; 33: 676-689.e673.
- Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response
to immunotherapy. Science (New York, N.Y.). 2017; 355.
- Ben-David U, Amon A. Context is everything: aneuploidy in cancer.
Nature reviews. Genetics. 2020; 21: 44-62.
- Lamberti G, Spurr LF, Li Y, et al. Clinicopathological and genomic
correlates of programmed cell death ligand 1 (PD-L1) expression in
nonsquamous non-small-cell lung cancer. Annals of oncology : official journal of the European Society for Medical Oncology. 2020;
31: 807-814.
- Spurr LF, Martinez CA, Kang W, et al. Highly aneuploidy non-small cell lung cancer shows enhanced responsiveness to
concurrent radiation and immune checkpoint blockade. Nature
cancer. 2022; 3: 1498-1512.
- Spurr LF, Weichselbaum RR, Pitroda SP. Tumor aneuploidy predicts survival following immunotherapy across multiple
cancers. Nature genetics. 2022; 54: 1782-1785.
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational
load predicts survival after immunotherapy across multiple cancer
types. Nature genetics. 2019; 51: 202-206.
- Salah Z, Itzhaki E, Aqeilan RI. The ubiquitin E3 ligase ITCH enhances
breast tumor progression by inhibiting the Hippo tumor suppressor pathway. Oncotarget. 2014; 5: 10886-10900.
- Chang L, Shen L, Zhou H, et al. ITCH nuclear translocation and H1.2
polyubiquitination negatively regulate the DNA damage response.
Nucleic acids research. 2019; 47: 824-842.
- Salah Z, Cohen S, Itzhaki E, Aqeilan RI. NEDD4 E3 ligase inhibits the
activity of the Hippo pathway by targeting LATS1 for degradation.
Cell cycle (Georgetown, Tex.). 2013; 12: 3817-3823.
- Yeung B, Ho KC, Yang X. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PloS one.
2013; 8: e61027.
- Kulis M, Esteller M. DNA methylation and cancer. Advances in genetics. 2010; 70: 27-56.